CorShrink on Toeplitz matrices
Kushal K Dey
1/28/2018
library(MASS)
library(glasso)
library(CorShrink)
library(corpcor)
DM_toeplitz = function(n,P){
library("MASS")
index1=sort(sample(seq(1:n),(n/2)))
index2=seq(1:n)[-index1]
Sigmatp=function(P){
a=array(0,dim=c(P,P))
for(i in 1:P){
for(j in 1:P){
a[i,j]=max(1-0.1*(abs(i-j)),0)
}
}
return(a)
}
Sigma = Sigmatp(P)
data = mvrnorm(n,rep(0,P),Sigma)
Xtest = data[index2,]
Xtrain = data[index1,]
Omega = solve(Sigma)
return(list(Xtrain = Xtrain, Xtest = Xtest, Sigma = Sigma))
}
n <- 10
P <- 200
ll <- DM_toeplitz(n=n, P=P)
data <- rbind(ll$Xtrain, ll$Xtest)
Sigma <- ll$Sigma
corSigma <- cov2cor(Sigma)
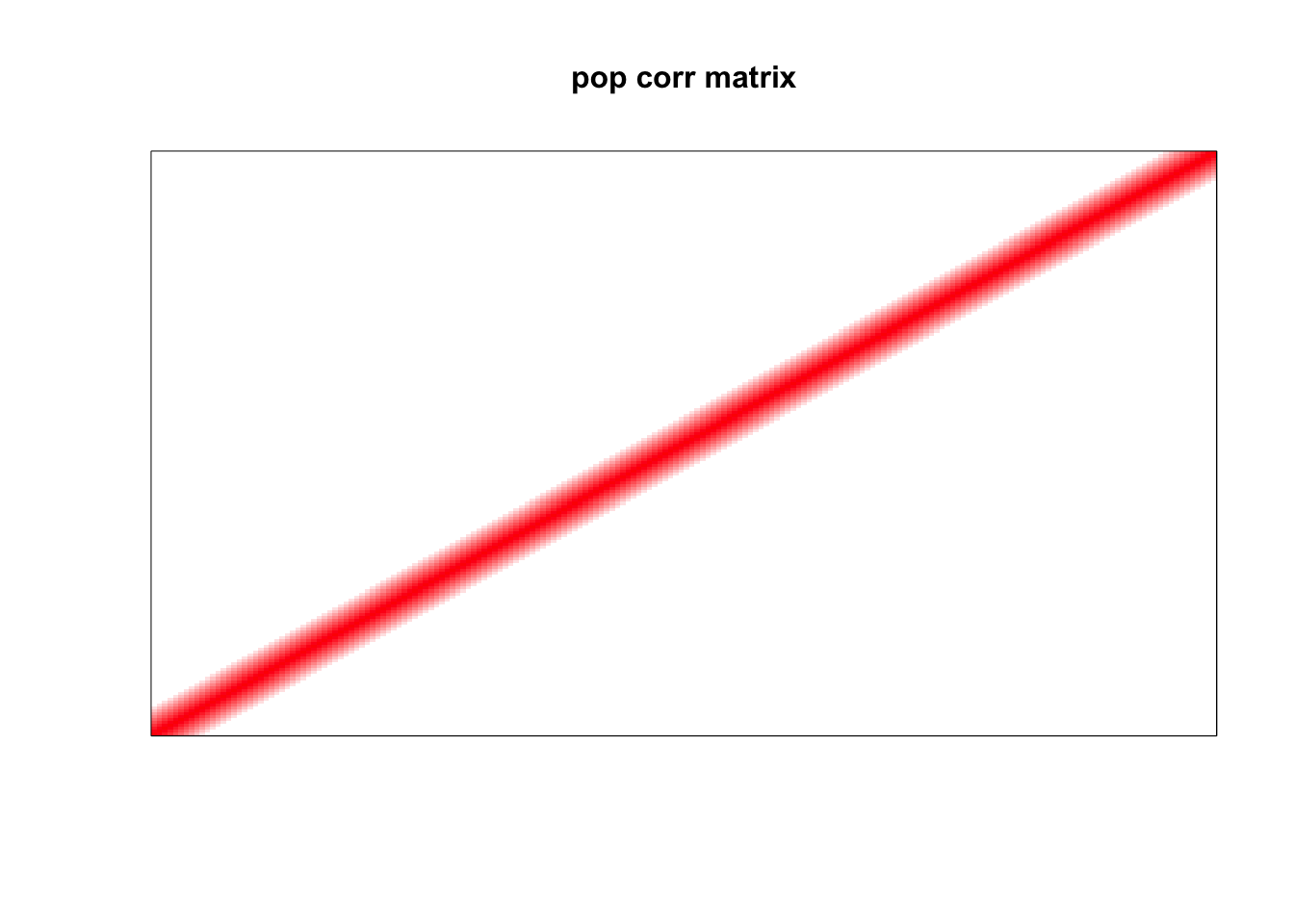
Population correlation matrix
col=c(rep(rgb(0,1,0), 1),rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)), rep(rgb(1,0,0),1))
image(as.matrix(corSigma),
col=col, main=paste0("pop corr matrix"), cex.main=1,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
Population precision matrix
col=c(rep(rgb(0,1,0), 1),rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)), rep(rgb(1,0,0),1))
image(solve(as.matrix(corSigma)),
col=col, main=paste0("pop precision matrix"), cex.main=1,
xaxt = "n", yaxt = "n")

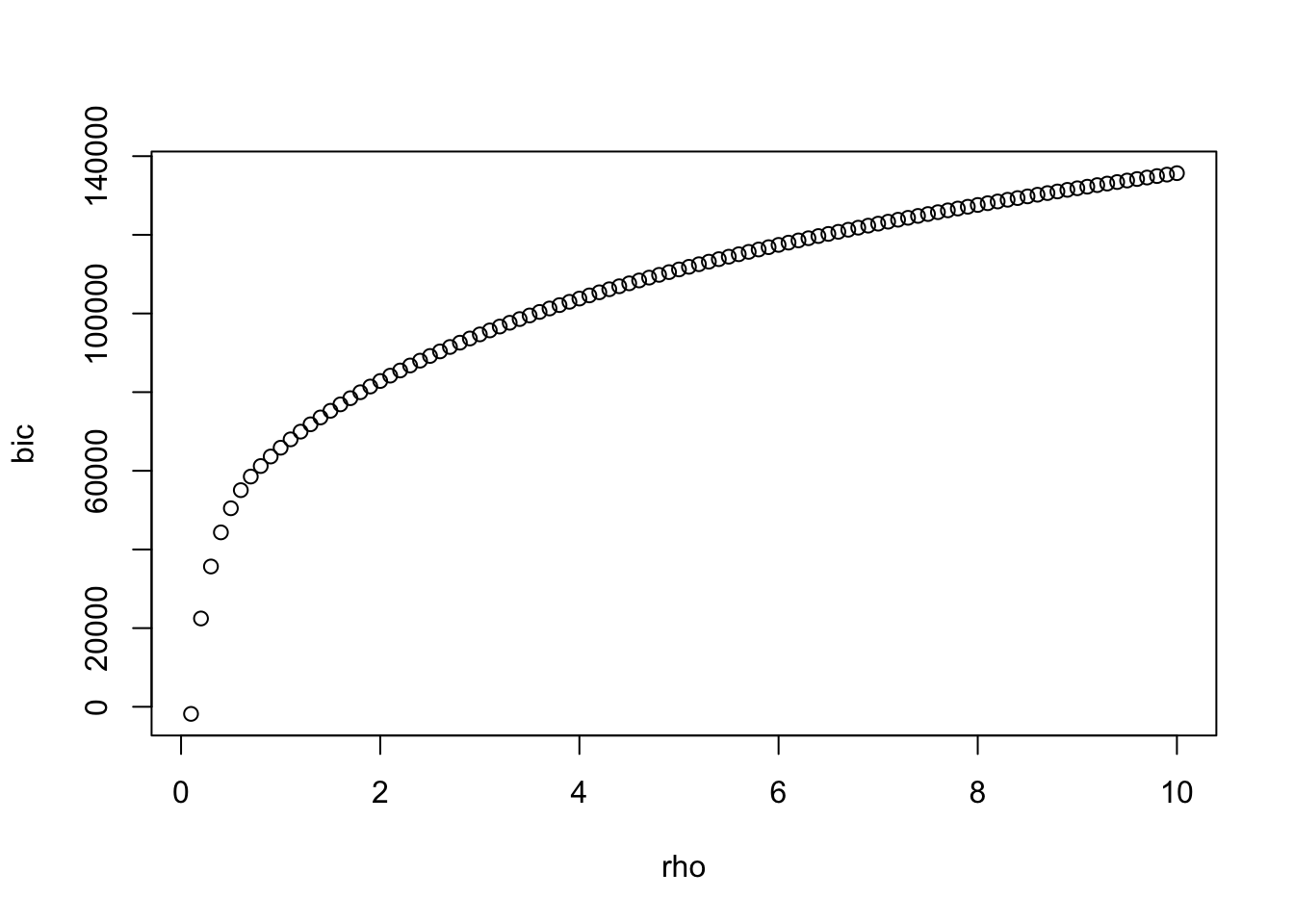
n = 10, p=200
nr <- 100
rho <- seq(0.1,10,length=nr)
bic <- rho
S <- cov(data)
for(j in 1:nr){
a <- glasso::glasso(S,rho[j])
p_off_d <- sum(a$wi!=0 & col(S)<row(S))
bic[j] <- -2*(a$loglik) + p_off_d*log(n)
}
best <- which.min(bic)
plot(rho,bic)
a <- glasso::glasso(S,rho[best])
## Strimmer Shafer
strimmer_sample <- corpcor::cor.shrink(data)
## Estimating optimal shrinkage intensity lambda (correlation matrix): 0.7407
## CorShrink
cov_sample_ML <- CorShrinkData(data, sd_boot = FALSE,
ash.control = list())
col=c(rep(rgb(0,1,0), 1),rev(rgb(seq(1,0,length=10000),1,seq(1,0,length=10000))),
rgb(1,seq(1,0,length=10000),seq(1,0,length=10000)), rep(rgb(1,0,0),1))
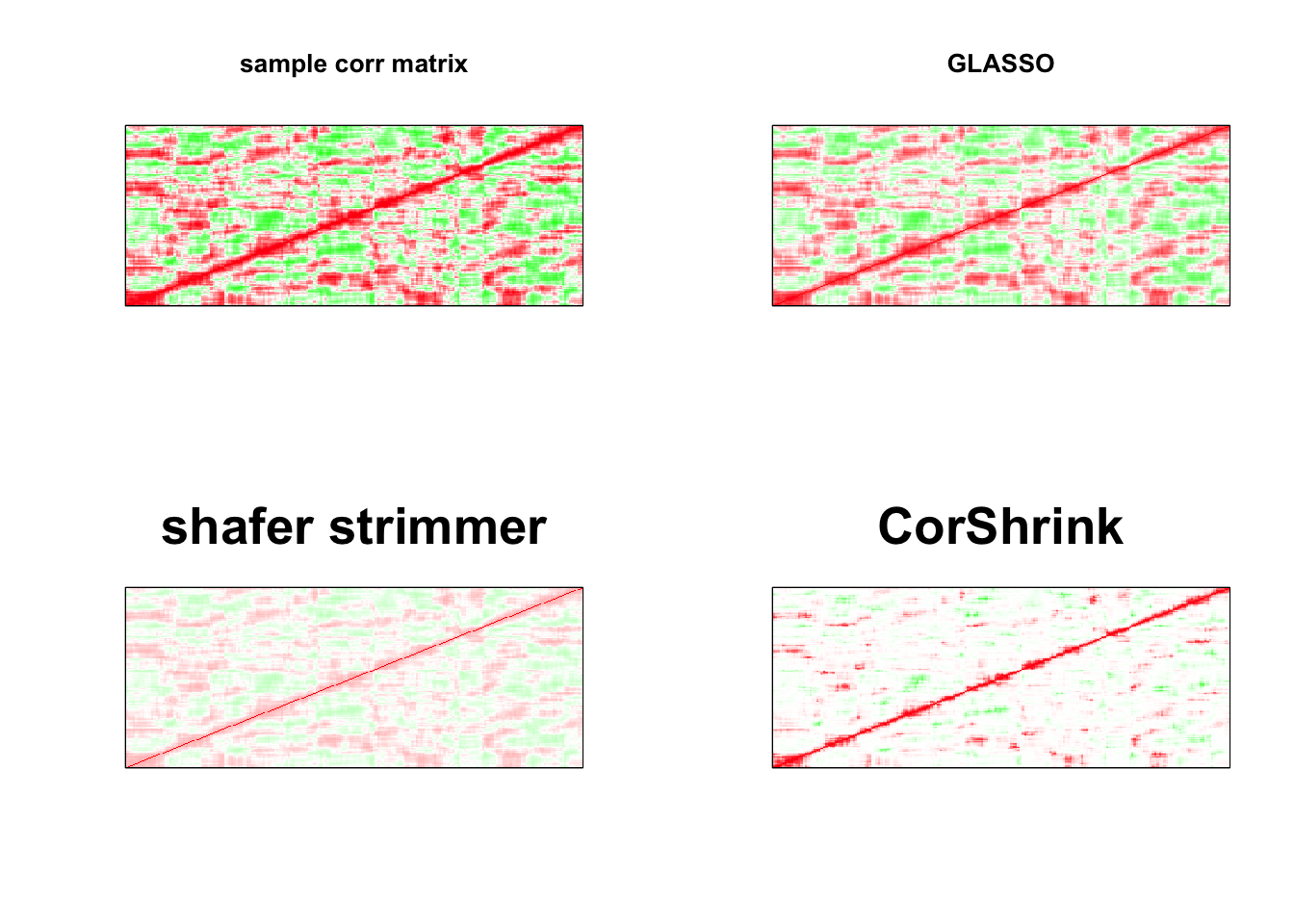
par(mfrow = c(2,2))
image(as.matrix(cov2cor(S)),
col=col, main=paste0("sample corr matrix"), cex.main=1,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
image(as.matrix(cov2cor(a$w)),
col=col, main=paste0("GLASSO"), cex.main=1,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
image(strimmer_sample,
col=col, main=paste0("shafer strimmer"), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
image(as.matrix(cov_sample_ML$cor),
col=col, main=paste0("CorShrink"), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
n = 25, p=200
n <- 25
P <- 200
ll <- DM_toeplitz(n=n, P=P)
data <- rbind(ll$Xtrain, ll$Xtest)
Sigma <- ll$Sigma
corSigma <- cov2cor(Sigma)
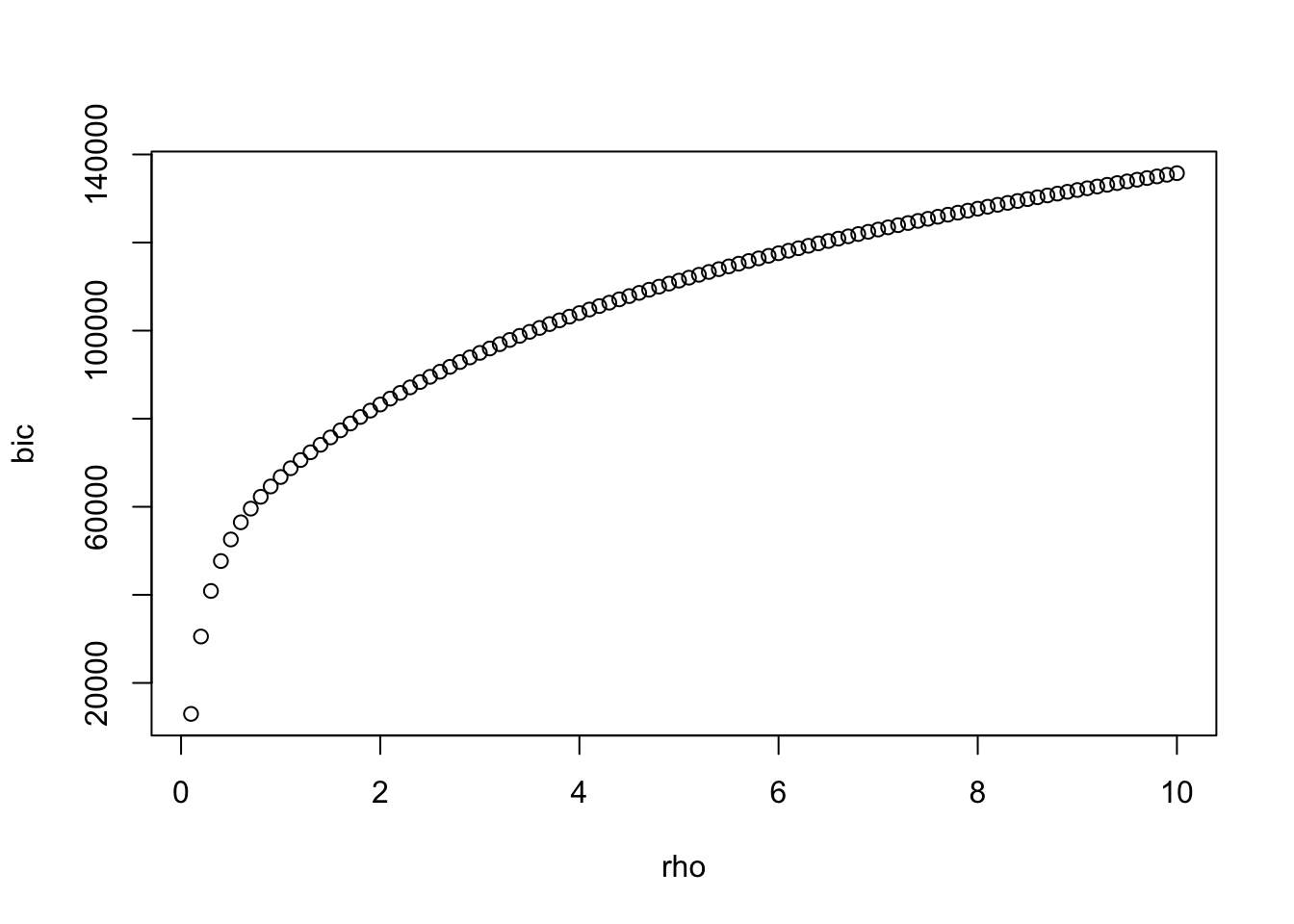
nr <- 100
rho <- seq(0.1,10,length=nr)
bic <- rho
S <- cov(data)
for(j in 1:nr){
a <- glasso::glasso(S,rho[j])
p_off_d <- sum(a$wi!=0 & col(S)<row(S))
bic[j] <- -2*(a$loglik) + p_off_d*log(n)
}
best <- which.min(bic)
plot(rho,bic)
a <- glasso::glasso(S,rho[best])
## Strimmer Shafer
strimmer_sample <- corpcor::cor.shrink(data)
## Estimating optimal shrinkage intensity lambda (correlation matrix): 0.5703
## CorShrink
cov_sample_ML <- CorShrinkData(data, sd_boot = FALSE,
ash.control = list())
col=c(rep(rgb(0,1,0), 1),rev(rgb(seq(1,0,length=10000),1,seq(1,0,length=10000))),
rgb(1,seq(1,0,length=10000),seq(1,0,length=10000)), rep(rgb(1,0,0),1))
par(mfrow = c(2,2))
image(as.matrix(cov2cor(S)),
col=col, main=paste0("sample corr matrix"), cex.main=1,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
image(as.matrix(cov2cor(a$w)),
col=col, main=paste0("GLASSO"), cex.main=1,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
image(strimmer_sample,
col=col, main=paste0("shafer strimmer"), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
image(as.matrix(cov_sample_ML$cor),
col=col, main=paste0("CorShrink"), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))

n = 50, p=200
n <- 50
P <- 200
ll <- DM_toeplitz(n=n, P=P)
data <- rbind(ll$Xtrain, ll$Xtest)
Sigma <- ll$Sigma
corSigma <- cov2cor(Sigma)
nr <- 100
rho <- seq(0.1,10,length=nr)
bic <- rho
S <- cov(data)
for(j in 1:nr){
a <- glasso::glasso(S,rho[j])
p_off_d <- sum(a$wi!=0 & col(S)<row(S))
bic[j] <- -2*(a$loglik) + p_off_d*log(n)
}
best <- which.min(bic)
plot(rho,bic)
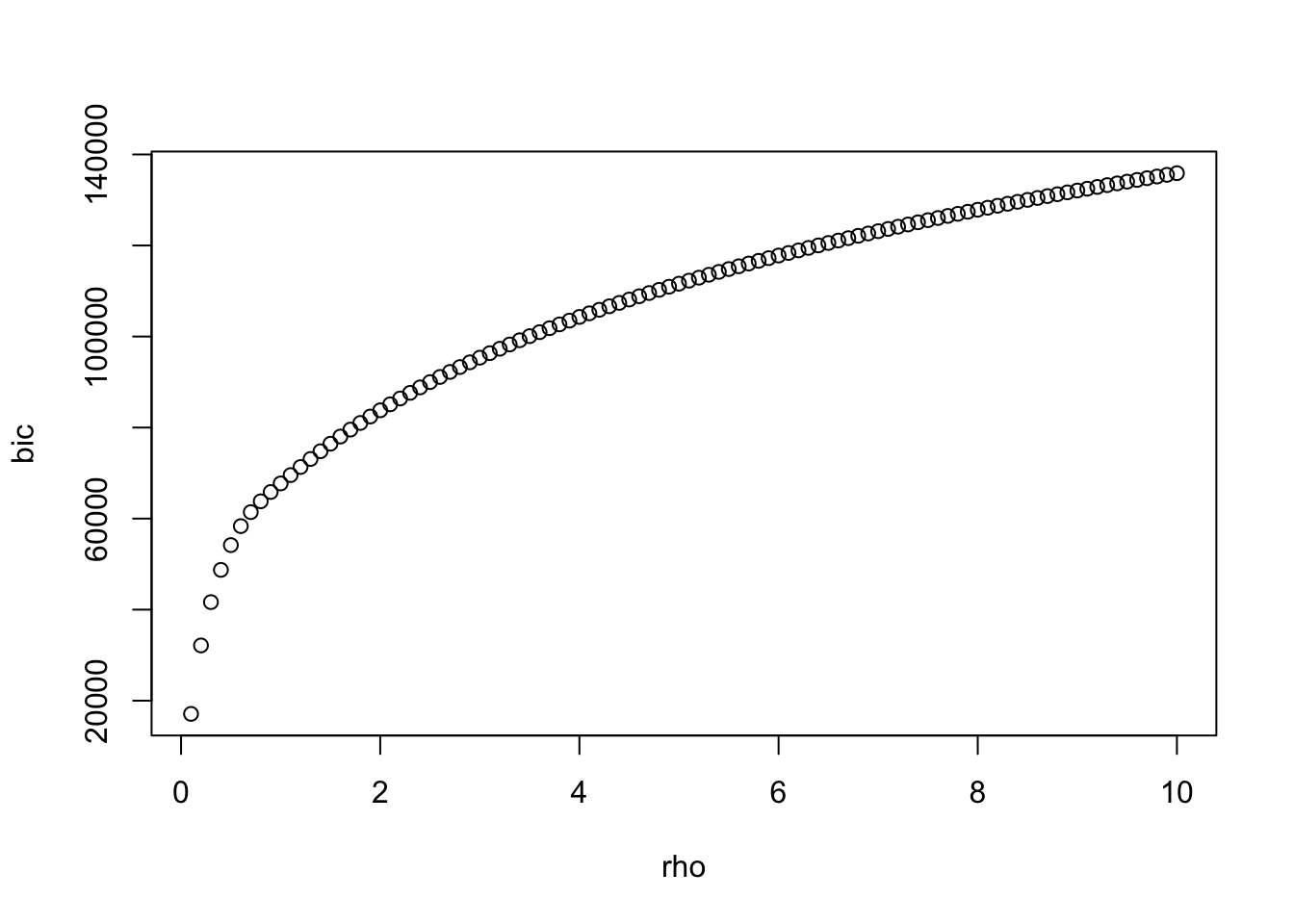
a <- glasso::glasso(S,rho[best])
## Strimmer Shafer
strimmer_sample <- corpcor::cor.shrink(data)
## Estimating optimal shrinkage intensity lambda (correlation matrix): 0.4244
## CorShrink
cov_sample_ML <- CorShrinkData(data, sd_boot = FALSE,
ash.control = list())
col=c(rep(rgb(0,1,0), 1),rev(rgb(seq(1,0,length=10000),1,seq(1,0,length=10000))),
rgb(1,seq(1,0,length=10000),seq(1,0,length=10000)), rep(rgb(1,0,0),1))
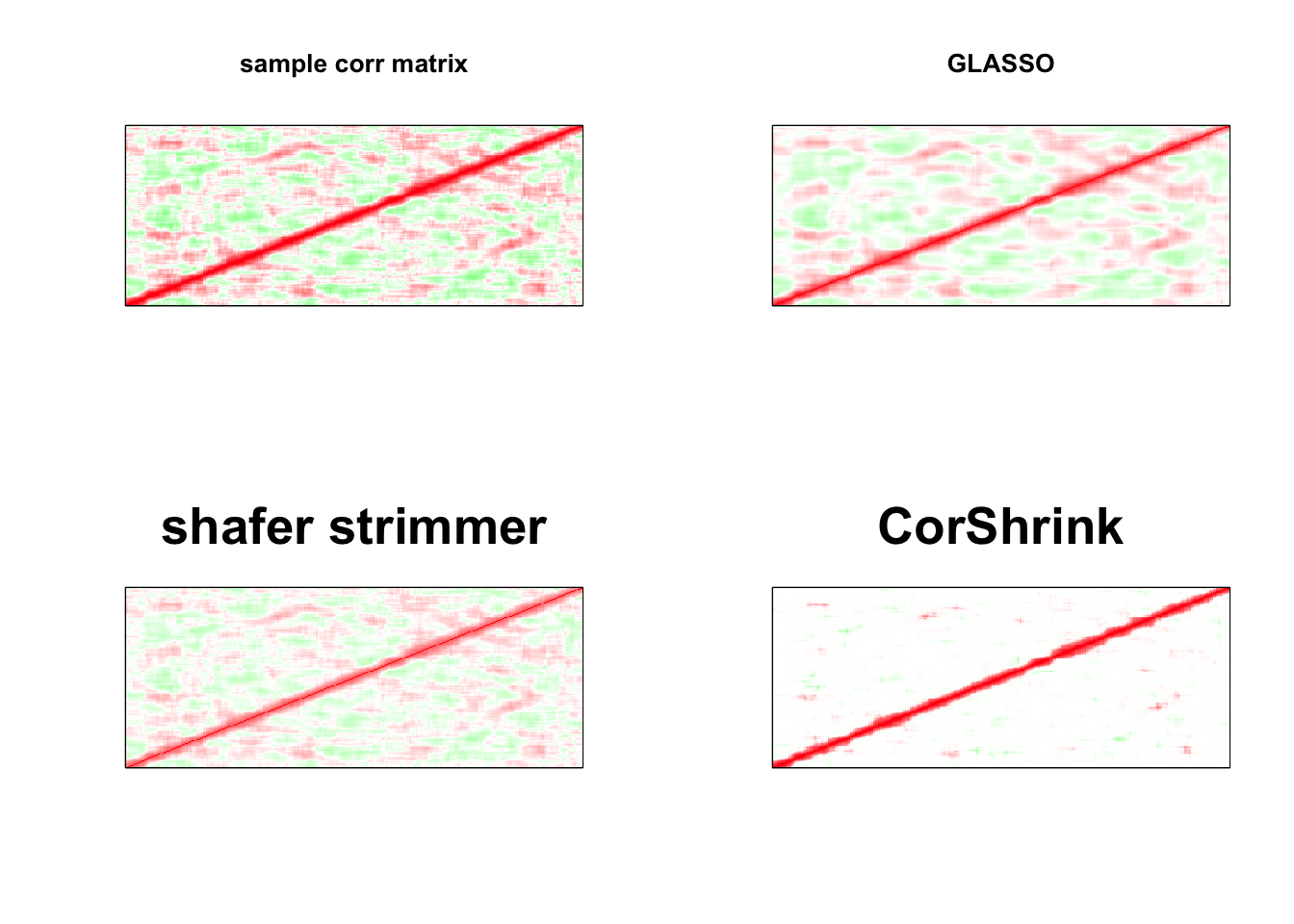
par(mfrow = c(2,2))
image(as.matrix(cov2cor(S)),
col=col, main=paste0("sample corr matrix"), cex.main=1,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
image(as.matrix(cov2cor(a$w)),
col=col, main=paste0("GLASSO"), cex.main=1,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
image(strimmer_sample,
col=col, main=paste0("shafer strimmer"), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
image(as.matrix(cov_sample_ML$cor),
col=col, main=paste0("CorShrink"), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
n = 75, p=200
n <- 75
P <- 200
ll <- DM_toeplitz(n=n, P=P)
data <- rbind(ll$Xtrain, ll$Xtest)
Sigma <- ll$Sigma
corSigma <- cov2cor(Sigma)
nr <- 100
rho <- seq(0.1,10,length=nr)
bic <- rho
S <- cov(data)
for(j in 1:nr){
a <- glasso::glasso(S,rho[j])
p_off_d <- sum(a$wi!=0 & col(S)<row(S))
bic[j] <- -2*(a$loglik) + p_off_d*log(n)
}
best <- which.min(bic)
plot(rho,bic)
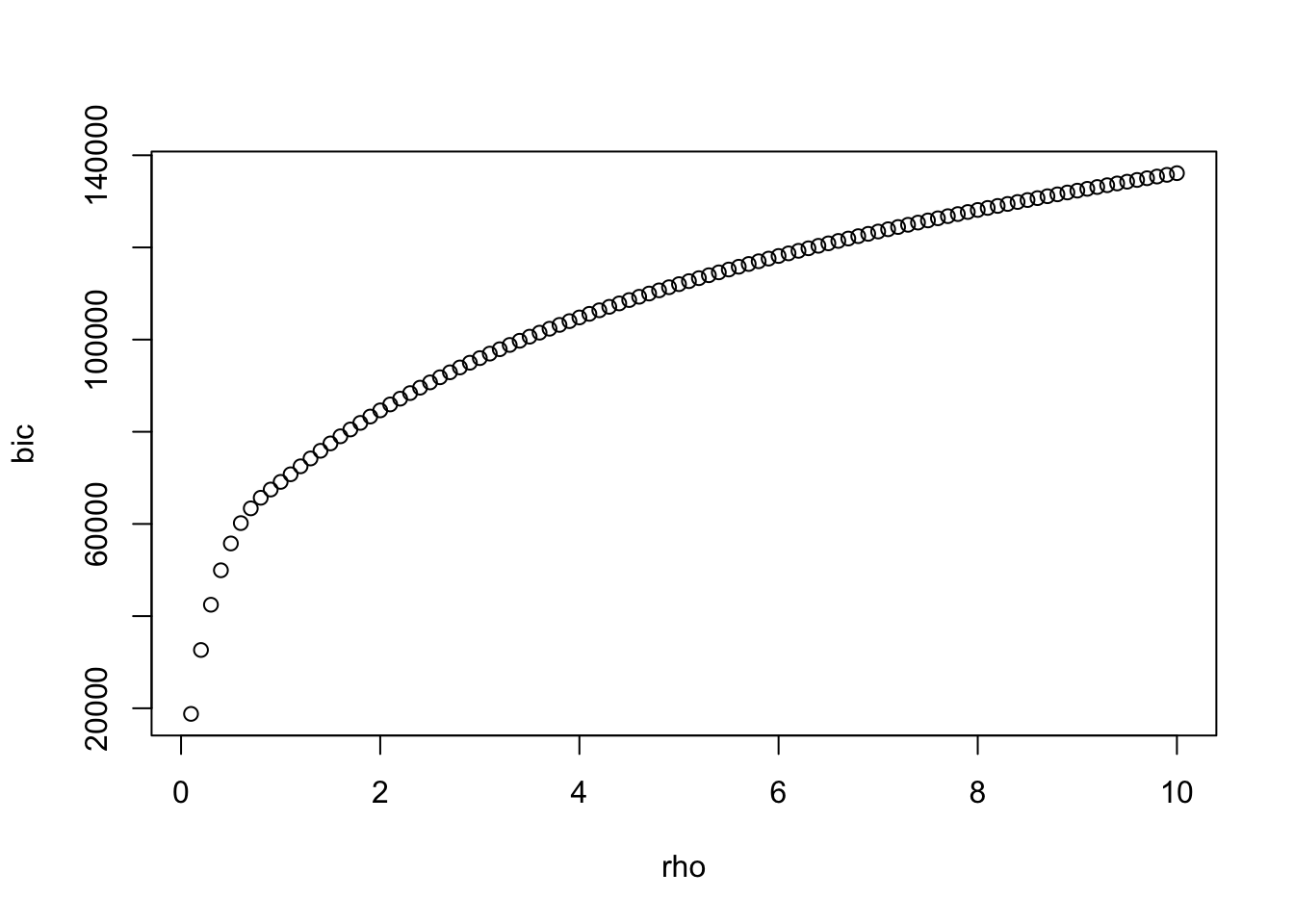
a <- glasso::glasso(S,rho[best])
## Strimmer Shafer
strimmer_sample <- corpcor::cor.shrink(data)
## Estimating optimal shrinkage intensity lambda (correlation matrix): 0.3216
## CorShrink
cov_sample_ML <- CorShrinkData(data, sd_boot = FALSE,
ash.control = list())
col=c(rep(rgb(0,1,0), 1),rev(rgb(seq(1,0,length=10000),1,seq(1,0,length=10000))),
rgb(1,seq(1,0,length=10000),seq(1,0,length=10000)), rep(rgb(1,0,0),1))
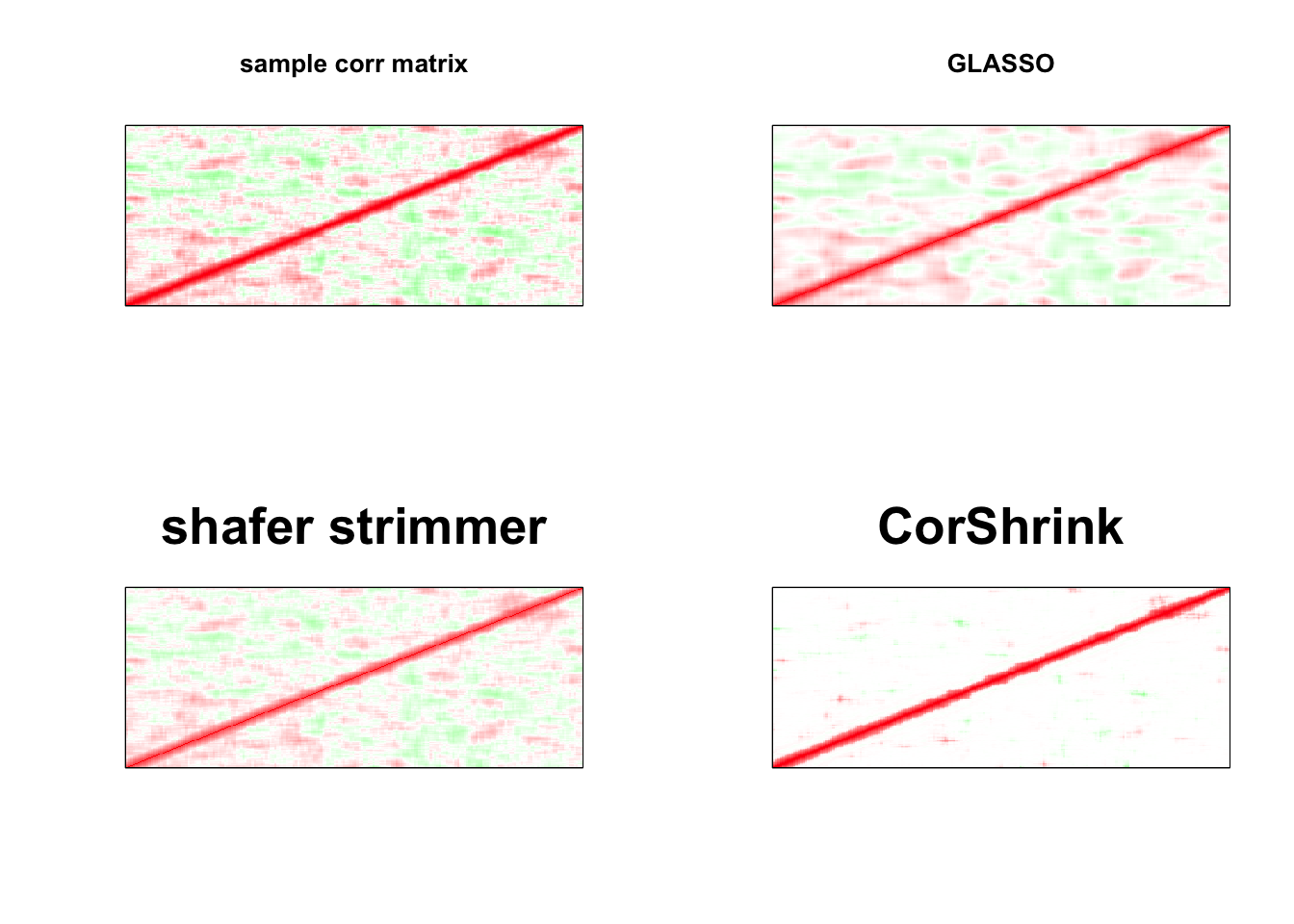
par(mfrow = c(2,2))
image(as.matrix(cov2cor(S)),
col=col, main=paste0("sample corr matrix"), cex.main=1,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
image(as.matrix(cov2cor(a$w)),
col=col, main=paste0("GLASSO"), cex.main=1,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
image(strimmer_sample,
col=col, main=paste0("shafer strimmer"), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
image(as.matrix(cov_sample_ML$cor),
col=col, main=paste0("CorShrink"), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
This R Markdown site was created with workflowr









