CorShrink on dsc simulated data (topelitz)
Kushal K Dey
6/22/2017
library(spcov)
library(CorShrink)
library(corpcor)
library(glasso)DM_toeplitz = function(n,P){
library("MASS")
index1=sort(sample(seq(1:n),(n/2)))
index2=seq(1:n)[-index1]
Sigmatp=function(P){
a=array(0,dim=c(P,P))
for(i in 1:P){
for(j in 1:P){
a[i,j]=max(1-0.1*(abs(i-j)),0)
}
}
return(a)
}
Sigma = Sigmatp(P)
data = mvrnorm(n,rep(0,P),Sigma)
Xtest = data[index2,]
Xtrain = data[index1,]
Omega = solve(Sigma)
return(list(Xtrain = Xtrain, Xtest = Xtest, Sigma = Sigma))
}n = 10, P=100
n <- 10
P <- 100
ll <- DM_toeplitz(n=n, P=P)
data <- rbind(ll$Xtrain, ll$Xtest)
Sigma <- ll$Sigma
corSigma <- cov2cor(Sigma)col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(corSigma),
col=col, main=paste0("sample corr: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))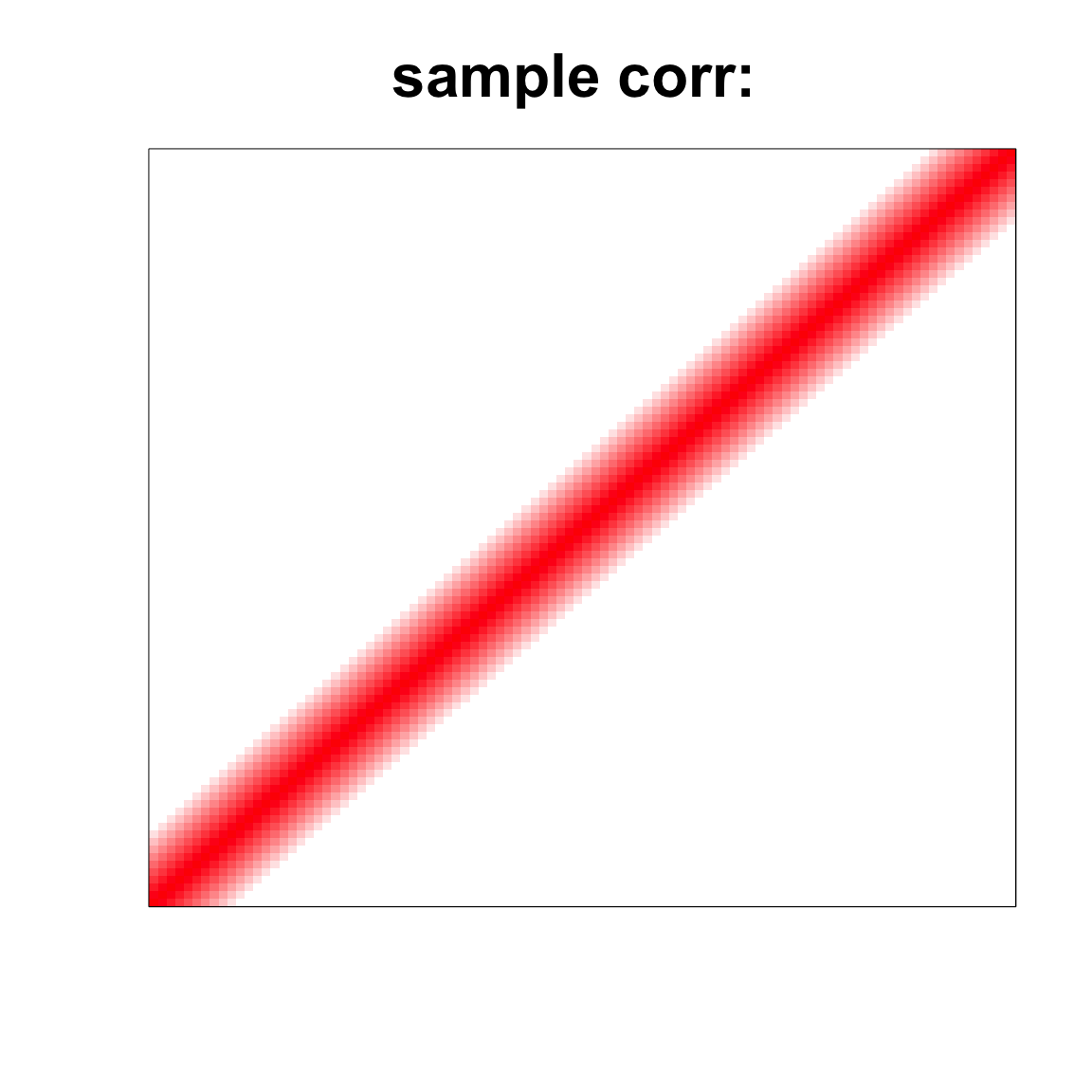
Pat <- matrix(1, P, P)
diag(Pat) <- 0lam <- 0.06
step.size <- 100
tol <- 1e-4
covmat <- cov(data)
mm <- spcov(Sigma=covmat+0.1*diag(1,P), S=covmat+0.1*diag(1,P), lambda=lam * Pat,step.size=step.size, n.inner.steps=200, thr.inner=0, tol.outer=tol, trace=1)## ---
## using Nesterov, backtracking line search
## ---
## objective: 92.93195
## step size given to GGDescent/Nesterov: 100
## objective: -44.75791 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 20
## step size given to GGDescent/Nesterov: 20
## objective: -45.91518 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 4
## step size given to GGDescent/Nesterov: 4
## objective: -45.92903 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.8
## step size given to GGDescent/Nesterov: 0.8
## objective: -45.93107 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.16
## step size given to GGDescent/Nesterov: 0.16
## objective: -45.93158 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.032
## step size given to GGDescent/Nesterov: 0.032
## objective: -45.93172 ( 200 iterations, max step size: 0.00128 )
## step size given to GGDescent/Nesterov: 0.032
## objective: -45.93176 ( 200 iterations, max step size: 0.00128 )
## MM converged in 7 steps!spcor_mat <- cov2cor(mm$Sigma)#devtools::install_github("kkdey/CorShrink")
#library(CorShrink)
#sessionInfo()
cov_mat <- cov(data);
system.time(strimmer_sample <- corpcor::cov.shrink(data))## Estimating optimal shrinkage intensity lambda.var (variance vector): 0.6672
##
## Estimating optimal shrinkage intensity lambda (correlation matrix): 0.6005## user system elapsed
## 0.022 0.001 0.024system.time(glasso_sample_005 <- glasso::glasso(cov_mat, rho = 0.05))## user system elapsed
## 0.442 0.003 0.453system.time(glasso_sample_05 <- glasso::glasso(cov_mat, rho = 0.5))## user system elapsed
## 0.085 0.001 0.087system.time(glasso_sample_1 <- glasso::glasso(cov_mat, rho = 1))## user system elapsed
## 0.004 0.000 0.003system.time(glasso_sample_10 <- glasso::glasso(cov_mat, rho = 10))## user system elapsed
## 0.001 0.000 0.001system.time(cov_sample_ML <- CorShrinkMatrix(cov2cor(cov_mat), nsamp = matrix(n, P, P), ash.control = list(mixcompdist = "normal", nullweight = 100)))## ash cor only and ash cor PD matrices are different## user system elapsed
## 2.144 0.044 2.230col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(cov_mat)),
col=col, main=paste0("sample corr: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))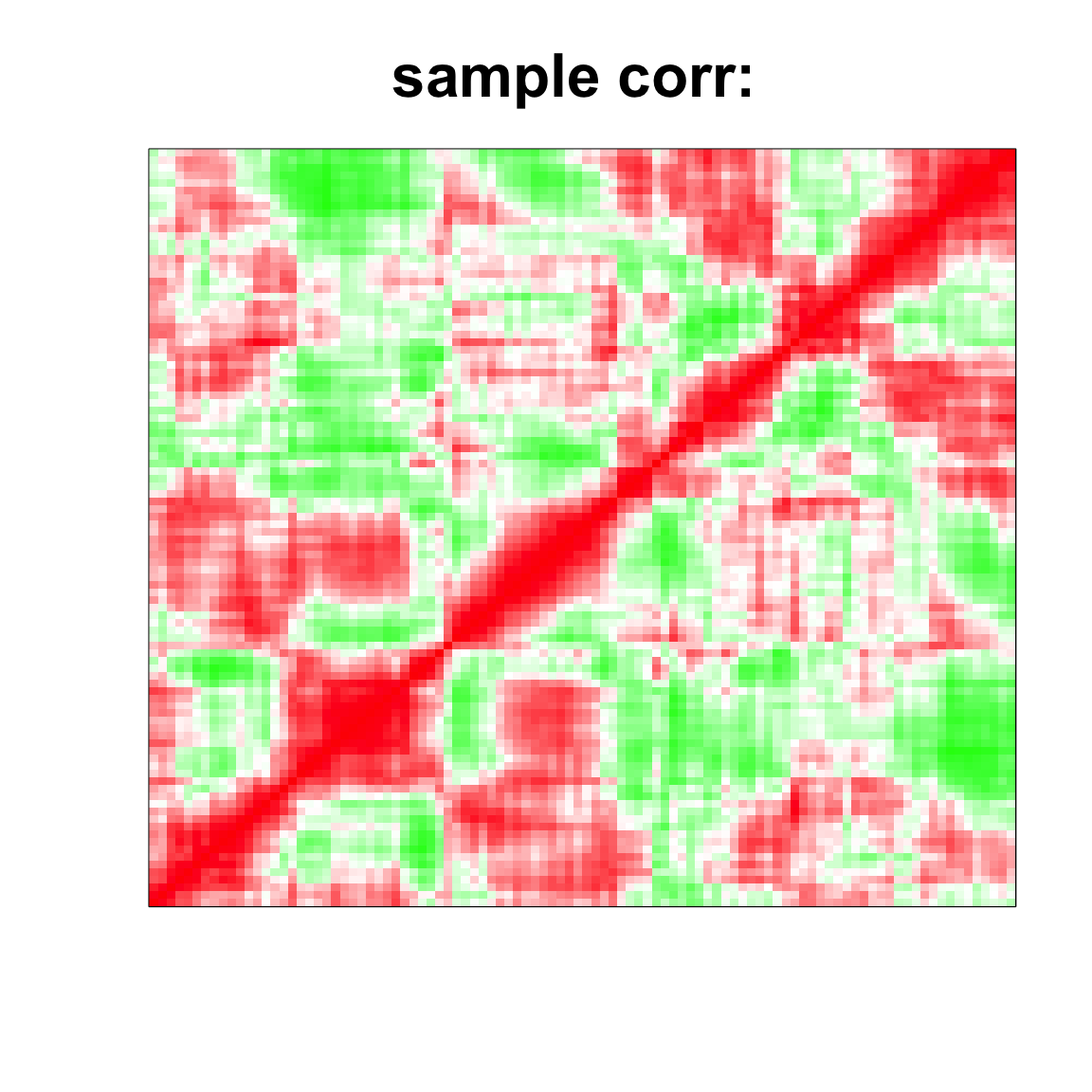
par(mfrow=c(2,2))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(strimmer_sample)),
col=col, main=paste0("shafer strimmer: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov_sample_ML$ash_cor_only),
col=col, main=paste0("corshrink: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(glasso_sample_005$w)),
col=col, main=paste0("glasso 0.05: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(glasso_sample_05$w)),
col=col, main=paste0("glasso 0.5: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))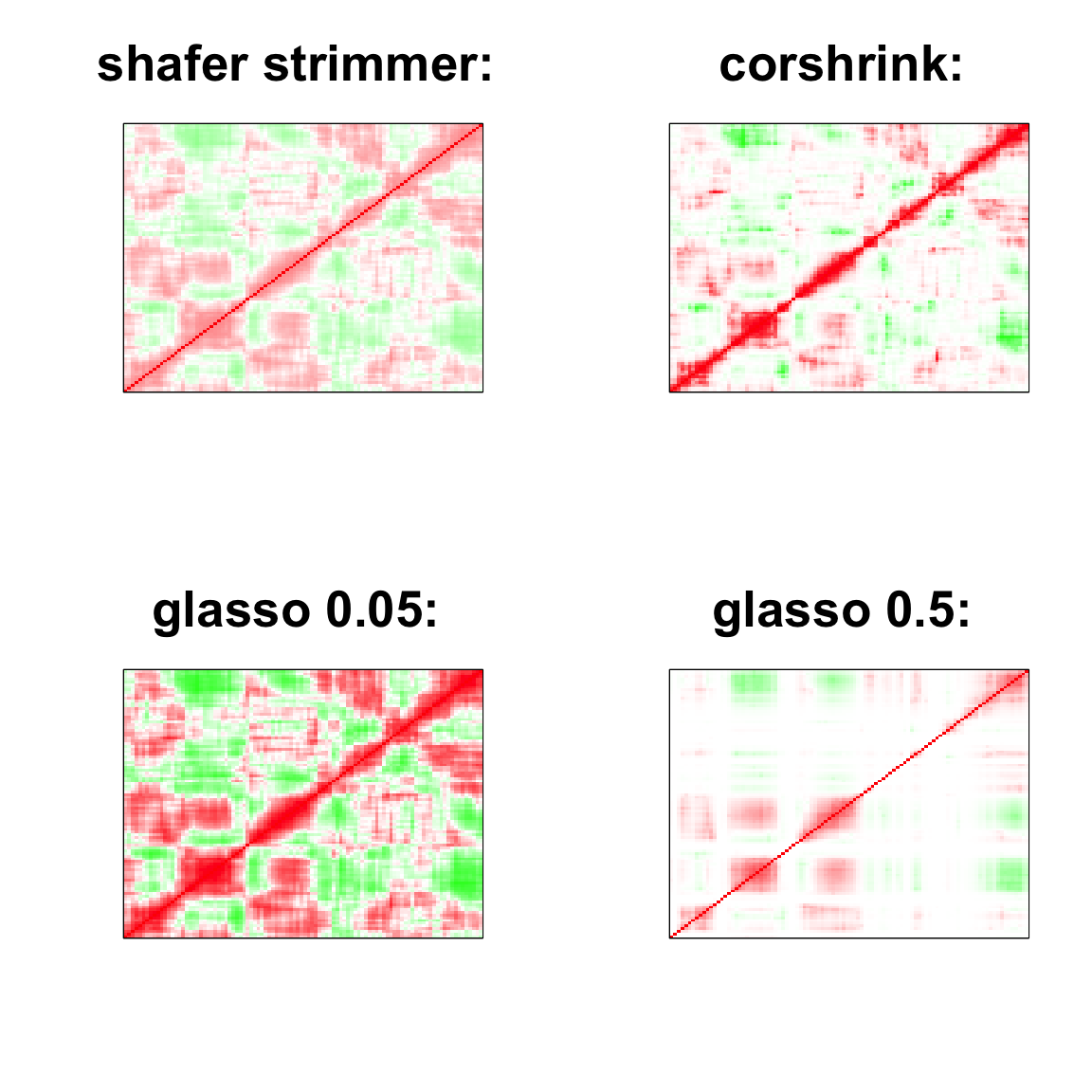
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(spcor_mat),
col=col, main=paste0("spcov:", expression(lambda), "=", lam), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))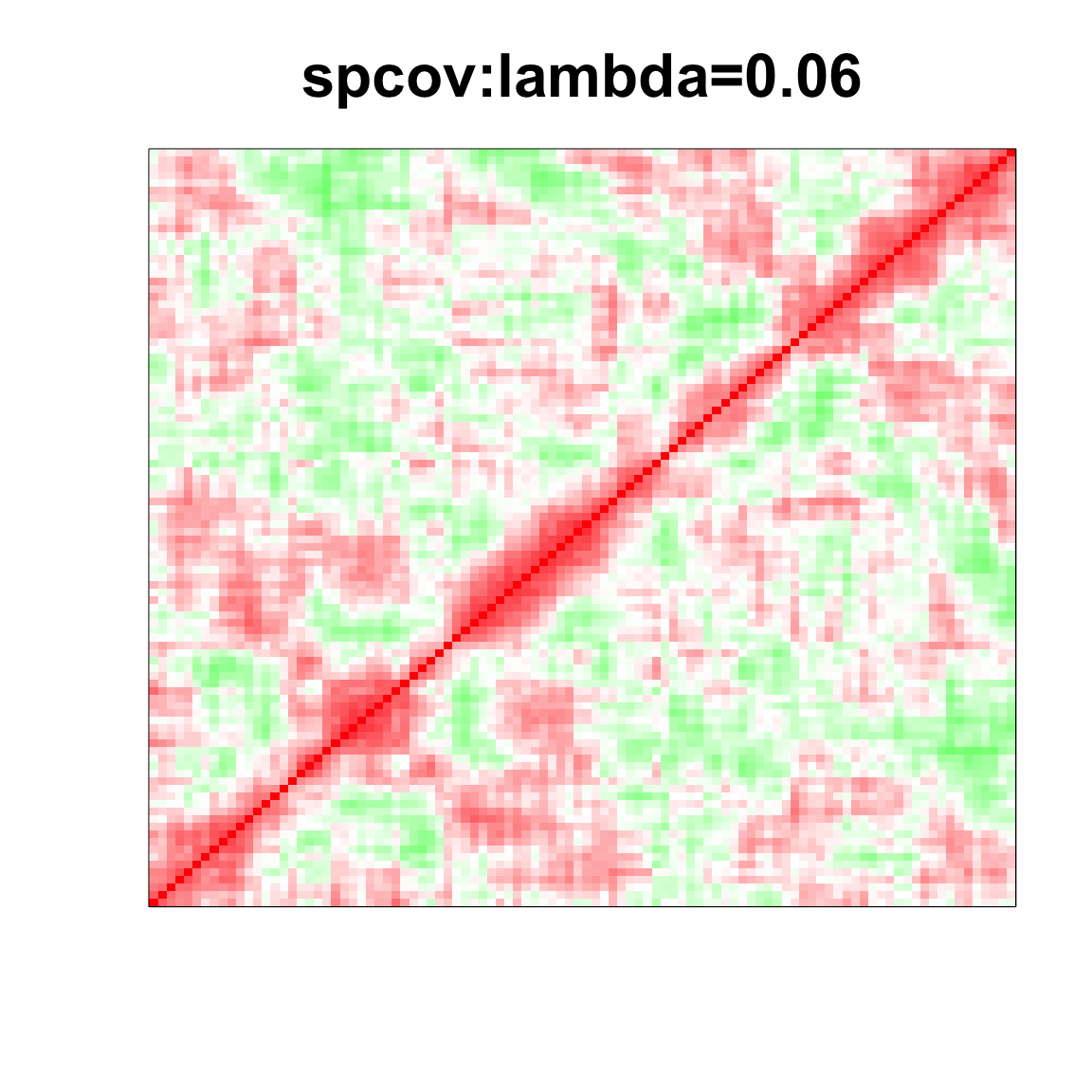
n = 50, P=100
n <- 50
P <- 100
ll <- DM_toeplitz(n=n, P=P)
data <- rbind(ll$Xtrain, ll$Xtest)
Sigma <- ll$Sigma
corSigma <- cov2cor(Sigma)Pat <- matrix(1, P, P)
diag(Pat) <- 0lam <- 0.06
step.size <- 100
tol <- 1e-4
covmat <- cov(data)
mm <- spcov(Sigma=covmat+0.1*diag(1,P), S=covmat+0.1*diag(1,P), lambda=lam * Pat,step.size=step.size, n.inner.steps=200, thr.inner=0, tol.outer=tol, trace=1)## ---
## using Nesterov, backtracking line search
## ---
## objective: 67.78083
## step size given to GGDescent/Nesterov: 100
## objective: -1.96839 ( 200 iterations, max step size: 0.032 )
## Reducing step size to 20
## step size given to GGDescent/Nesterov: 20
## objective: -2.570267 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 4
## step size given to GGDescent/Nesterov: 4
## objective: -2.620866 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 0.8
## step size given to GGDescent/Nesterov: 0.8
## objective: -2.634745 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 0.16
## step size given to GGDescent/Nesterov: 0.16
## objective: -2.640305 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 0.032
## step size given to GGDescent/Nesterov: 0.032
## objective: -2.642824 ( 200 iterations, max step size: 0.0064 )
## step size given to GGDescent/Nesterov: 0.032
## objective: -2.644047 ( 200 iterations, max step size: 0.0064 )
## step size given to GGDescent/Nesterov: 0.032
## objective: -2.64467 ( 200 iterations, max step size: 0.0064 )
## step size given to GGDescent/Nesterov: 0.032
## objective: -2.645002 ( 200 iterations, max step size: 0.0064 )
## step size given to GGDescent/Nesterov: 0.032
## objective: -2.645184 ( 200 iterations, max step size: 0.0064 )
## step size given to GGDescent/Nesterov: 0.032
## objective: -2.645287 ( 200 iterations, max step size: 0.0064 )
## step size given to GGDescent/Nesterov: 0.032
## objective: -2.645345 ( 200 iterations, max step size: 0.0064 )
## MM converged in 12 steps!spcor_mat <- cov2cor(mm$Sigma)#devtools::install_github("kkdey/CorShrink")
#library(CorShrink)
#sessionInfo()
cov_mat <- cov(data);
system.time(strimmer_sample <- corpcor::cov.shrink(data))## Estimating optimal shrinkage intensity lambda.var (variance vector): 1
##
## Estimating optimal shrinkage intensity lambda (correlation matrix): 0.2985## user system elapsed
## 0.078 0.000 0.078system.time(glasso_sample_005 <- glasso::glasso(cov_mat, rho = 0.05))## user system elapsed
## 0.253 0.000 0.255system.time(glasso_sample_05 <- glasso::glasso(cov_mat, rho = 0.5))## user system elapsed
## 0.045 0.001 0.046system.time(glasso_sample_1 <- glasso::glasso(cov_mat, rho = 1))## user system elapsed
## 0.002 0.000 0.002system.time(glasso_sample_10 <- glasso::glasso(cov_mat, rho = 10))## user system elapsed
## 0.001 0.000 0.002system.time(cov_sample_ML <- CorShrinkMatrix(cov2cor(cov_mat), nsamp = matrix(n, P, P), ash.control = list(mixcompdist = "normal", nullweight = 100)))## ash cor only and ash cor PD matrices are different## user system elapsed
## 1.709 0.058 1.774col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(cov_mat)),
col=col, main=paste0("sample corr: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))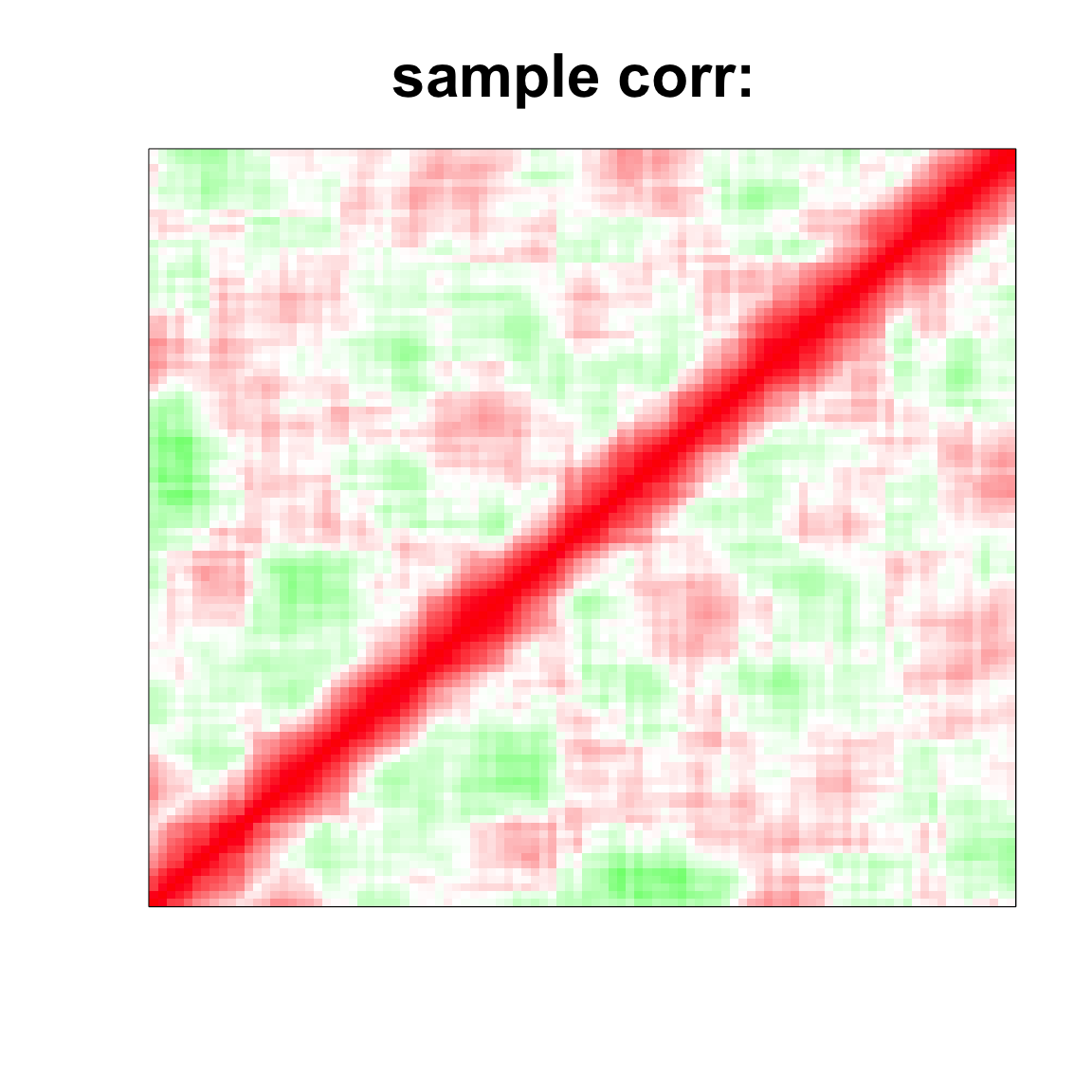
par(mfrow = c(2,2))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(strimmer_sample)),
col=col, main=paste0("shafer strimmer: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov_sample_ML$ash_cor_only),
col=col, main=paste0("corshrink: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(glasso_sample_005$w)),
col=col, main=paste0("glasso 0.05: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(glasso_sample_05$w)),
col=col, main=paste0("glasso 0.5: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))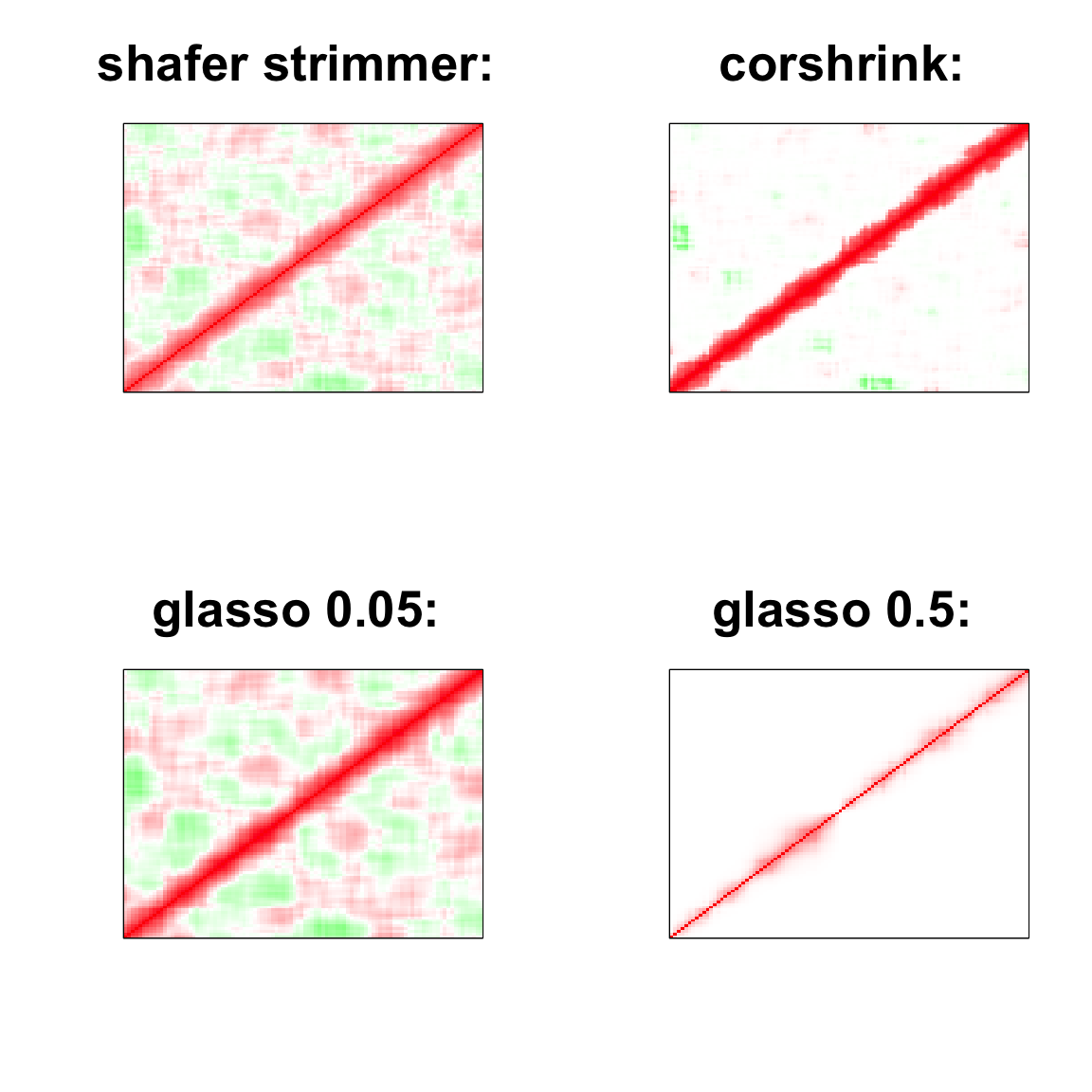
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(spcor_mat),
col=col, main=paste0("spcov:", expression(lambda), "=", lam), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))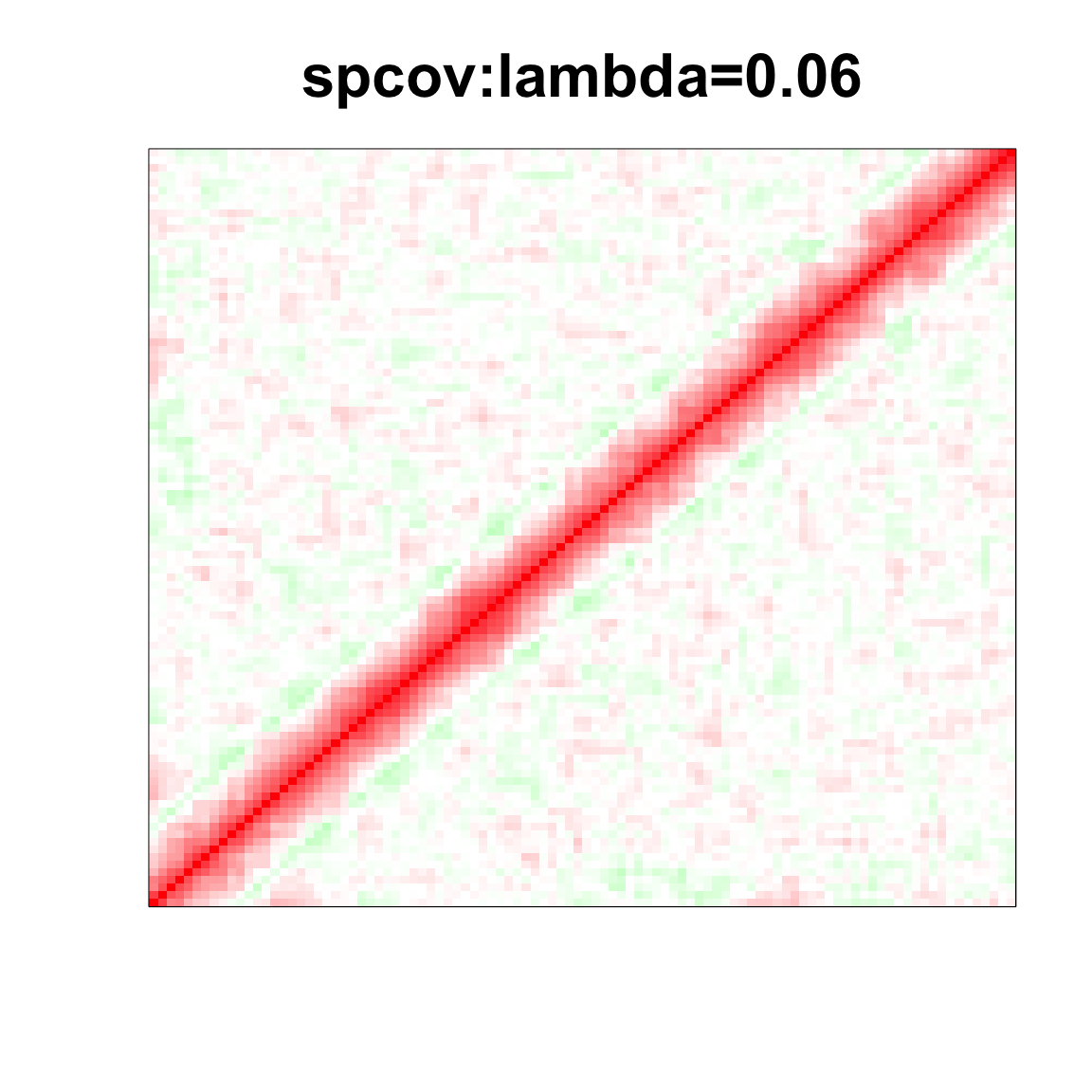
n = 5, P=100
n <- 5
P <- 100
ll <- DM_toeplitz(n=n, P=P)
data <- rbind(ll$Xtrain, ll$Xtest)
Sigma <- ll$Sigma
corSigma <- cov2cor(Sigma)Pat <- matrix(1, P, P)
diag(Pat) <- 0lam <- 0.06
step.size <- 100
tol <- 1e-4
covmat <- cov(data)
mm <- spcov(Sigma=covmat+0.1*diag(1,P), S=covmat+0.1*diag(1,P), lambda=lam * Pat,step.size=step.size, n.inner.steps=200, thr.inner=0, tol.outer=tol, trace=1)## ---
## using Nesterov, backtracking line search
## ---
## objective: 56.47488
## step size given to GGDescent/Nesterov: 100
## objective: -71.14374 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 20
## step size given to GGDescent/Nesterov: 20
## objective: -73.84421 ( 200 iterations, max step size: 0.032 )
## Reducing step size to 4
## step size given to GGDescent/Nesterov: 4
## objective: -73.85986 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.8
## step size given to GGDescent/Nesterov: 0.8
## objective: -73.86161 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.16
## step size given to GGDescent/Nesterov: 0.16
## objective: -73.86207 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.032
## step size given to GGDescent/Nesterov: 0.032
## objective: -73.8622 ( 200 iterations, max step size: 0.00128 )
## step size given to GGDescent/Nesterov: 0.032
## objective: -73.86223 ( 200 iterations, max step size: 0.00128 )
## MM converged in 7 steps!spcor_mat <- cov2cor(mm$Sigma)#devtools::install_github("kkdey/CorShrink")
#library(CorShrink)
#sessionInfo()
cov_mat <- cov(data);
system.time(strimmer_sample <- corpcor::cov.shrink(data))## Estimating optimal shrinkage intensity lambda.var (variance vector): 0.6363
##
## Estimating optimal shrinkage intensity lambda (correlation matrix): 0.6646## user system elapsed
## 0.009 0.000 0.009system.time(glasso_sample_005 <- glasso::glasso(cov_mat, rho = 0.05))## user system elapsed
## 0.566 0.002 0.570system.time(glasso_sample_05 <- glasso::glasso(cov_mat, rho = 0.5))## user system elapsed
## 0.034 0.001 0.034system.time(glasso_sample_1 <- glasso::glasso(cov_mat, rho = 1))## user system elapsed
## 0.003 0.000 0.002system.time(glasso_sample_10 <- glasso::glasso(cov_mat, rho = 10))## user system elapsed
## 0.002 0.000 0.002system.time(cov_sample_ML <- CorShrinkMatrix(cov2cor(cov_mat), nsamp = matrix(n, P, P), ash.control = list(mixcompdist = "normal", nullweight = 100)))## ash cor only and ash cor PD matrices are different## user system elapsed
## 2.815 0.072 2.900col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(cov_mat)),
col=col, main=paste0("sample corr: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))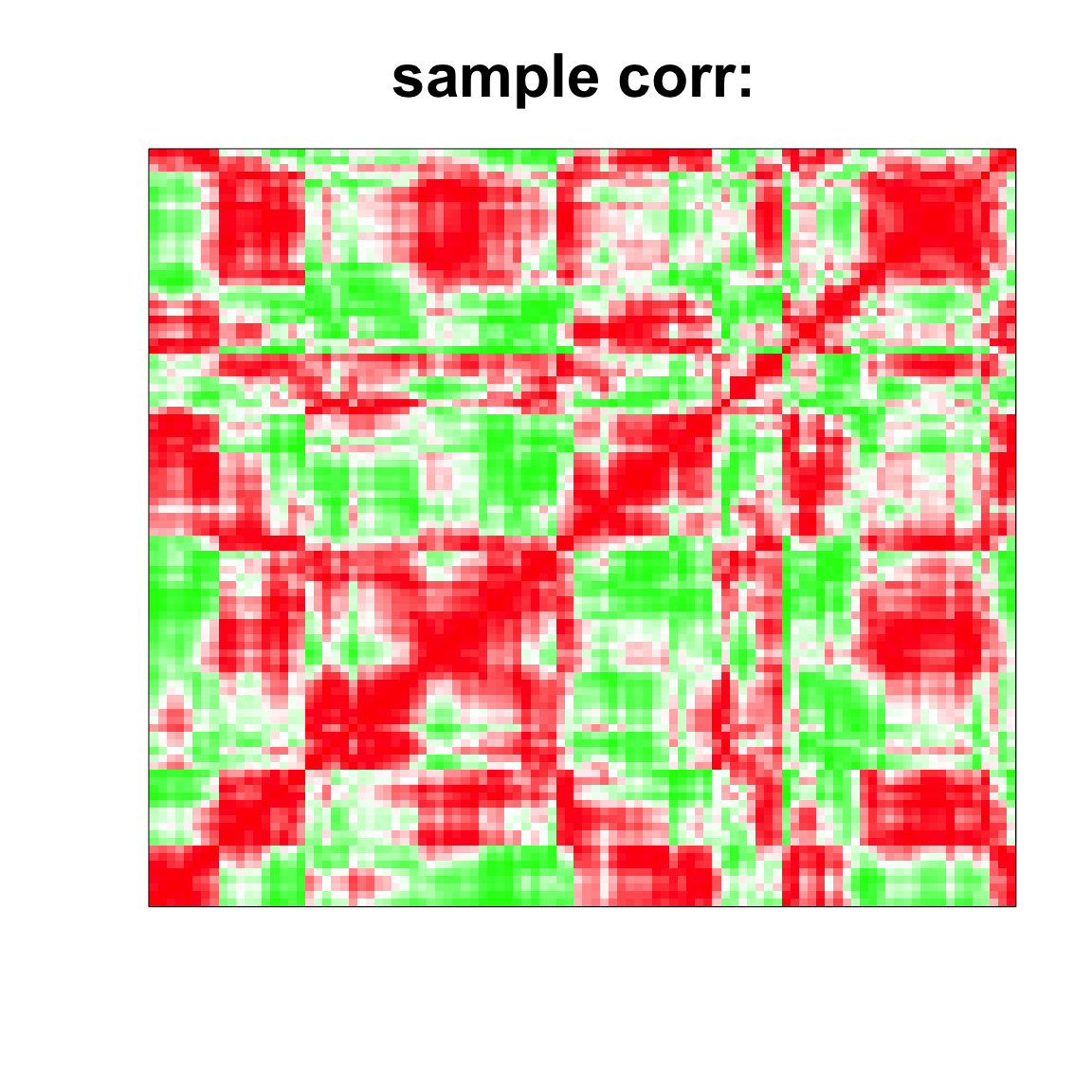
par(mfrow = c(2,2))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(strimmer_sample)),
col=col, main=paste0("shafer strimmer: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov_sample_ML$ash_cor_only),
col=col, main=paste0("corshrink: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(glasso_sample_005$w)),
col=col, main=paste0("glasso 0.05: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(glasso_sample_05$w)),
col=col, main=paste0("glasso 0.5: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))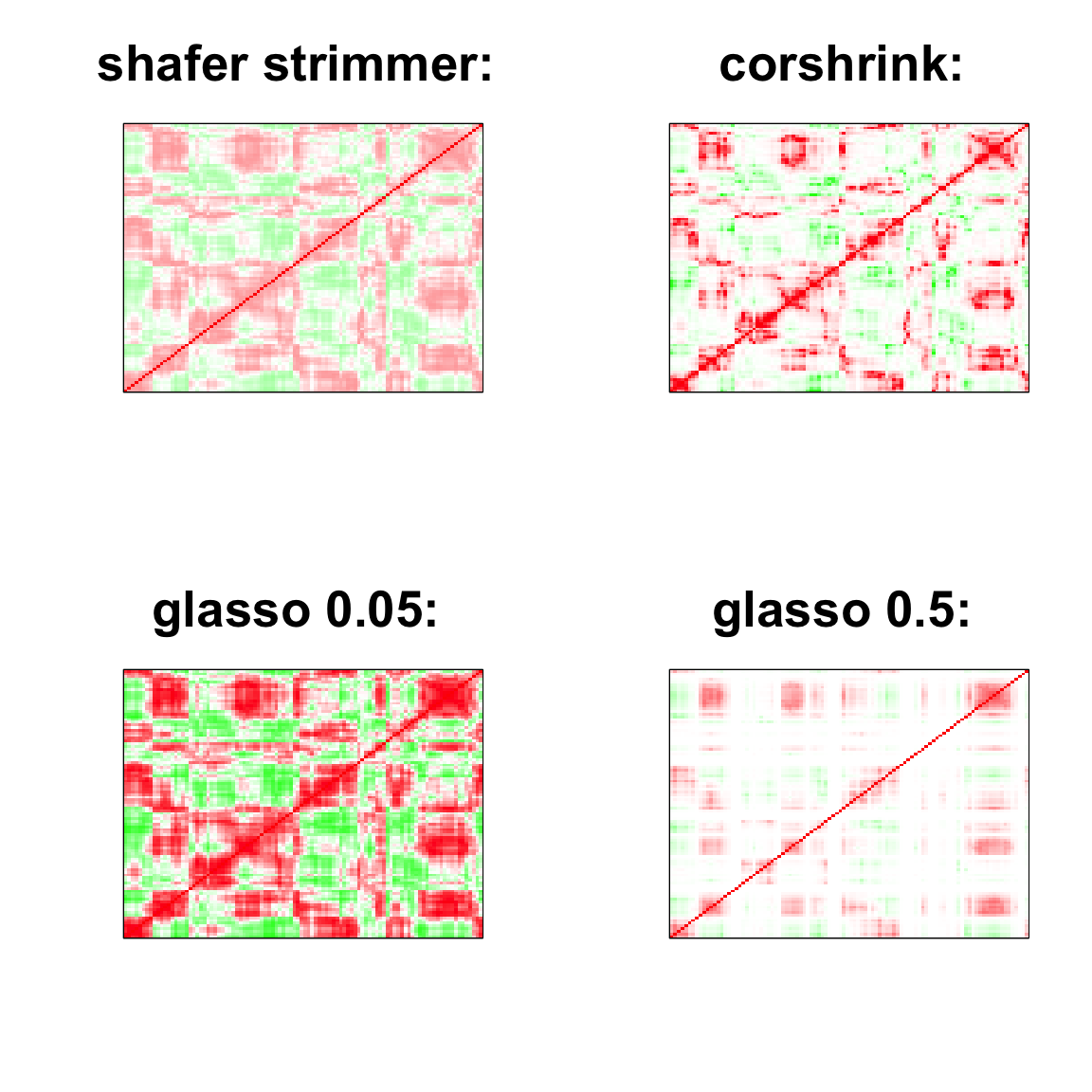
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(spcor_mat),
col=col, main=paste0("spcov:", expression(lambda), "=", lam), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))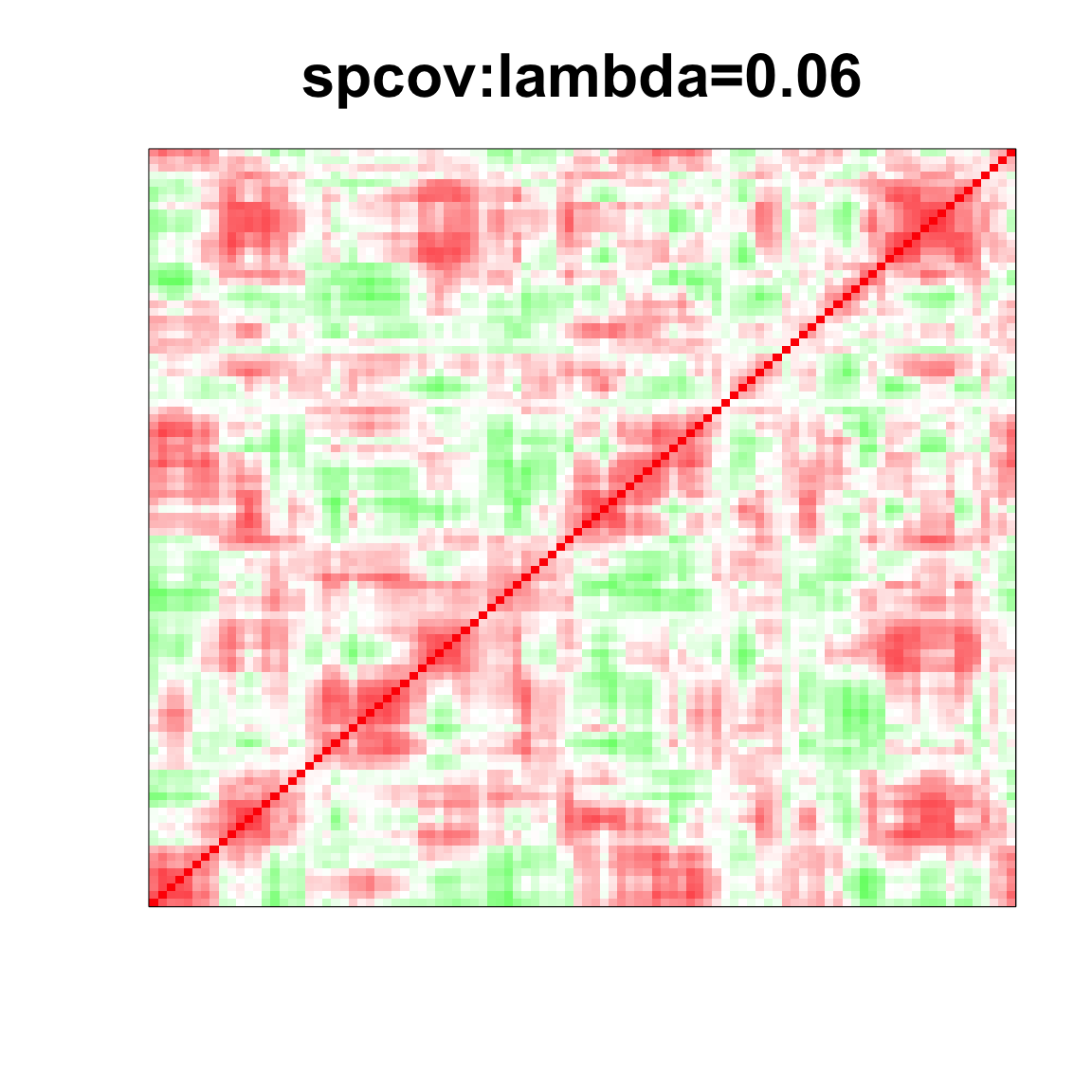
This R Markdown site was created with workflowr