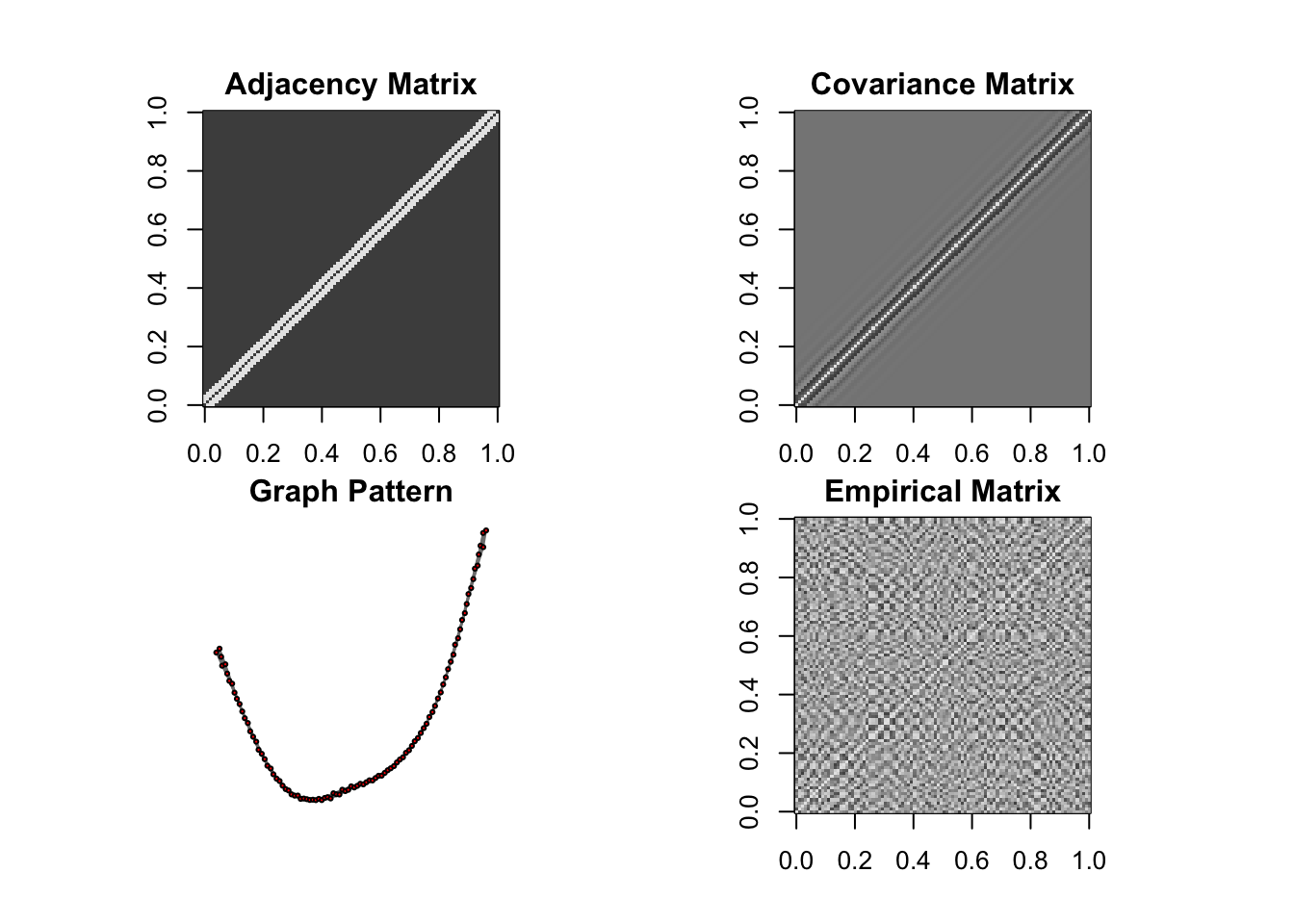
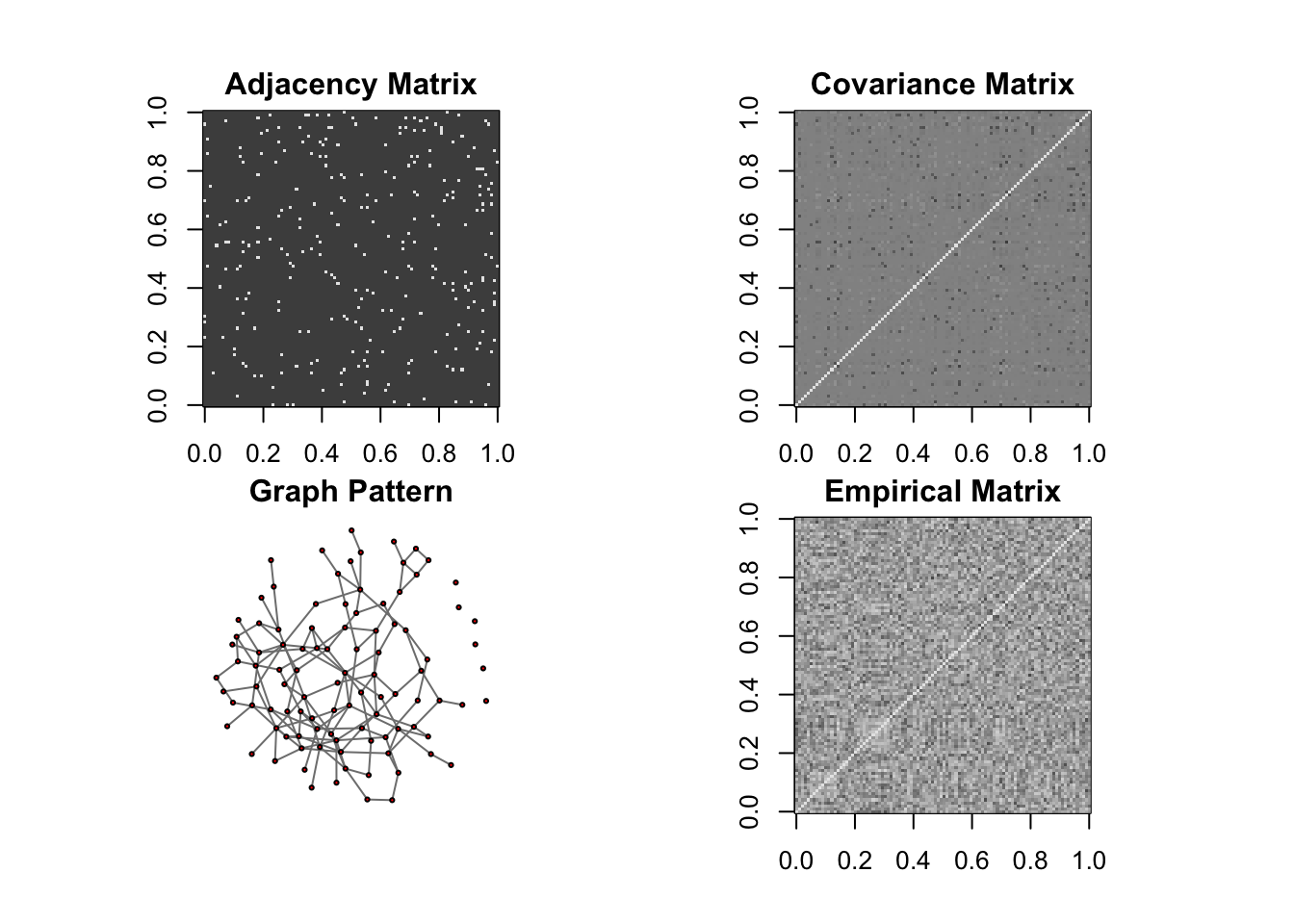
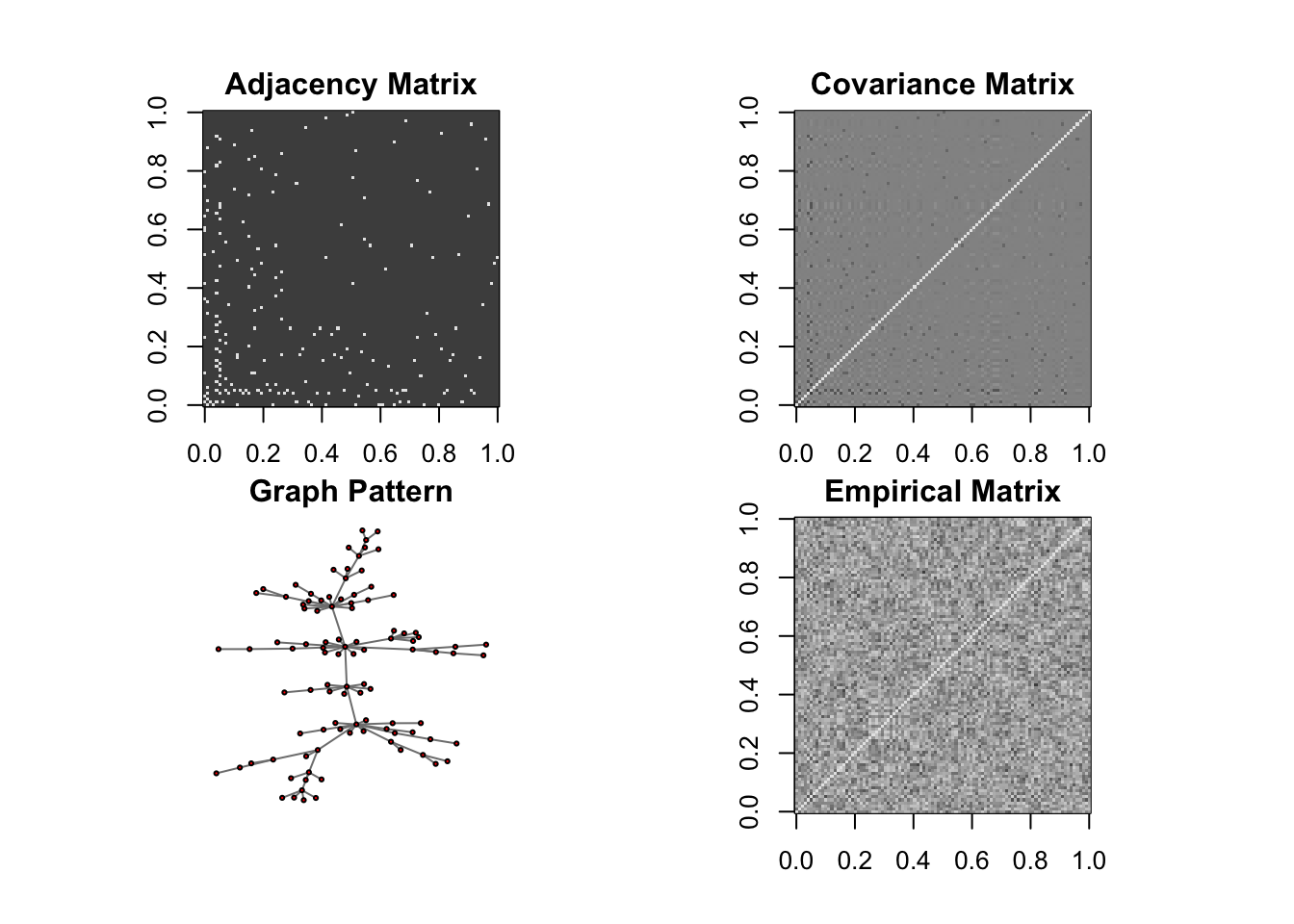
DM graph (band)
n = 10, P=100
n <- 10
P <- 100
ll <- DM_graph(n,P,g_type = "band",para = 3)
## Generating data from the multivariate normal distribution with the band graph structure....
## done.
data <- rbind(ll$Xtrain, ll$Xtest)
Sigma <- ll$Sigma
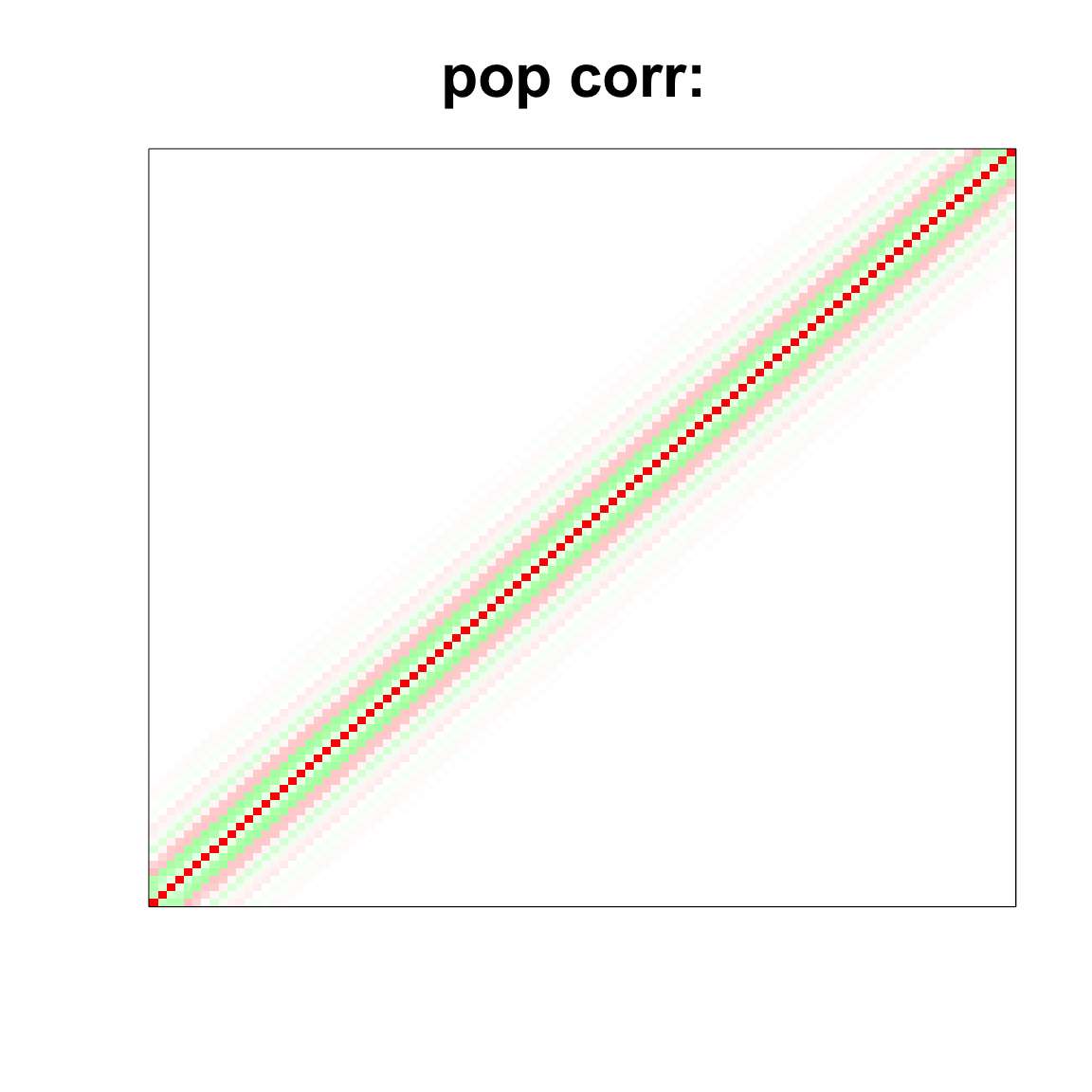
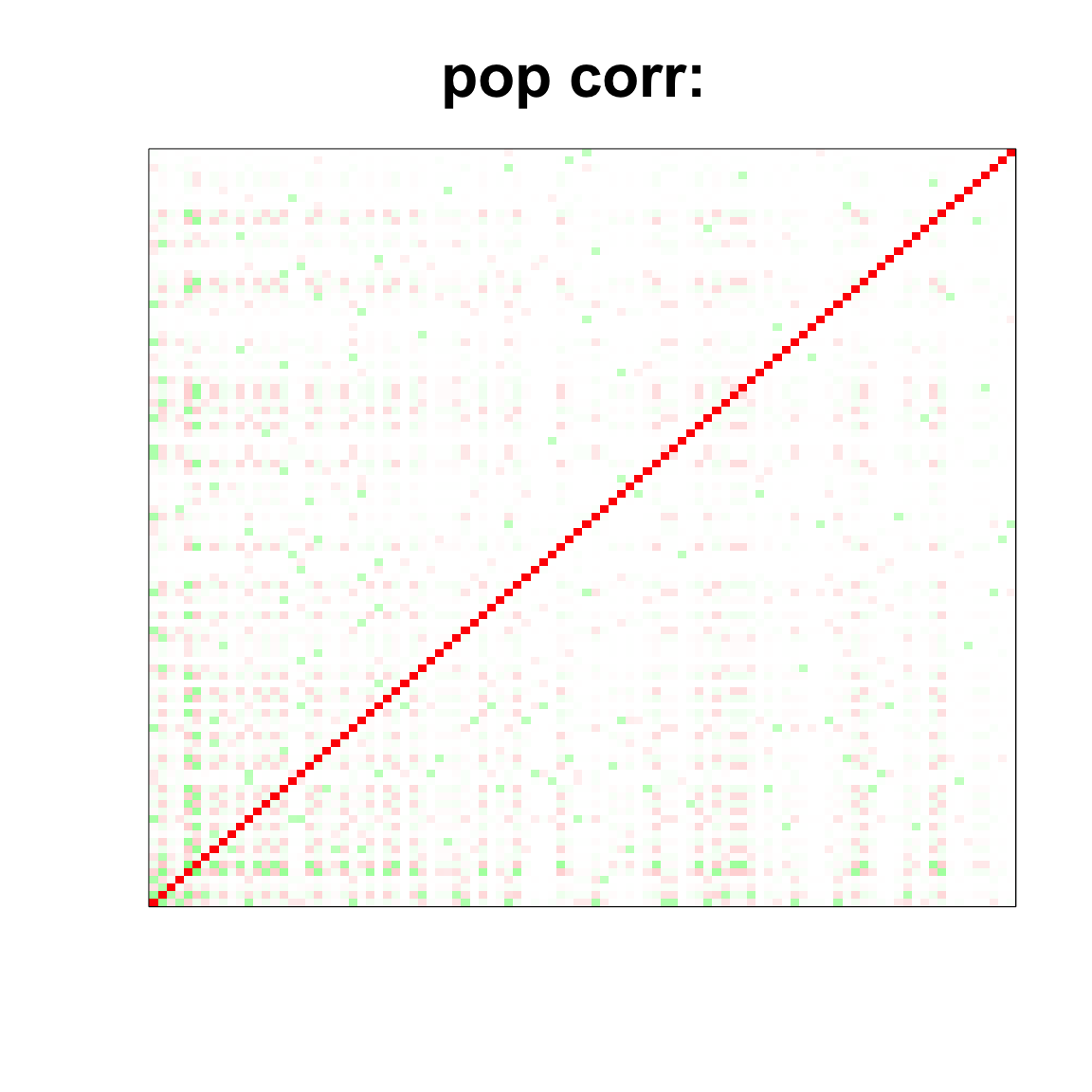
corSigma <- cov2cor(Sigma)
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
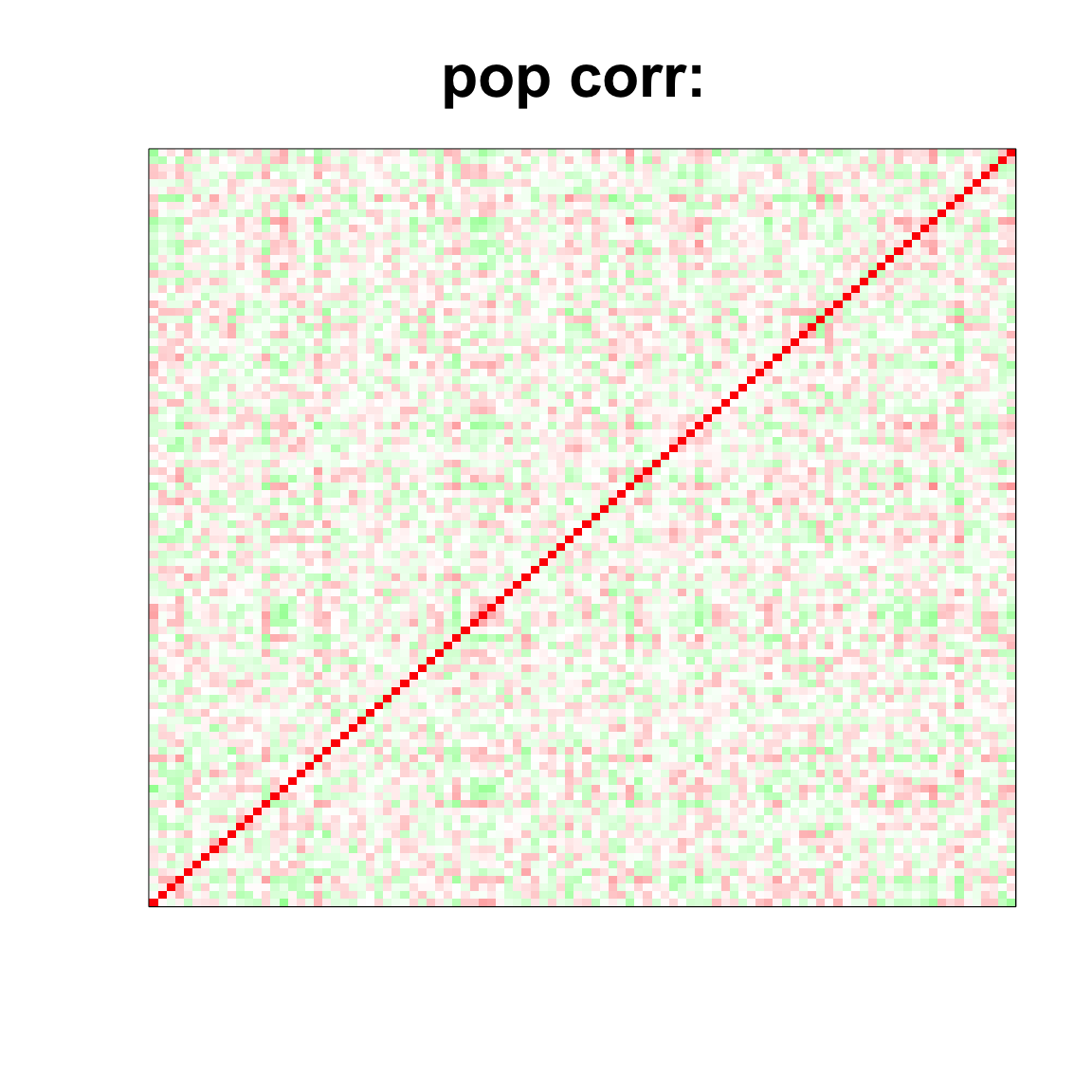
image(as.matrix(corSigma),
col=col, main=paste0("pop corr: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
Pat <- matrix(1, P, P)
diag(Pat) <- 0
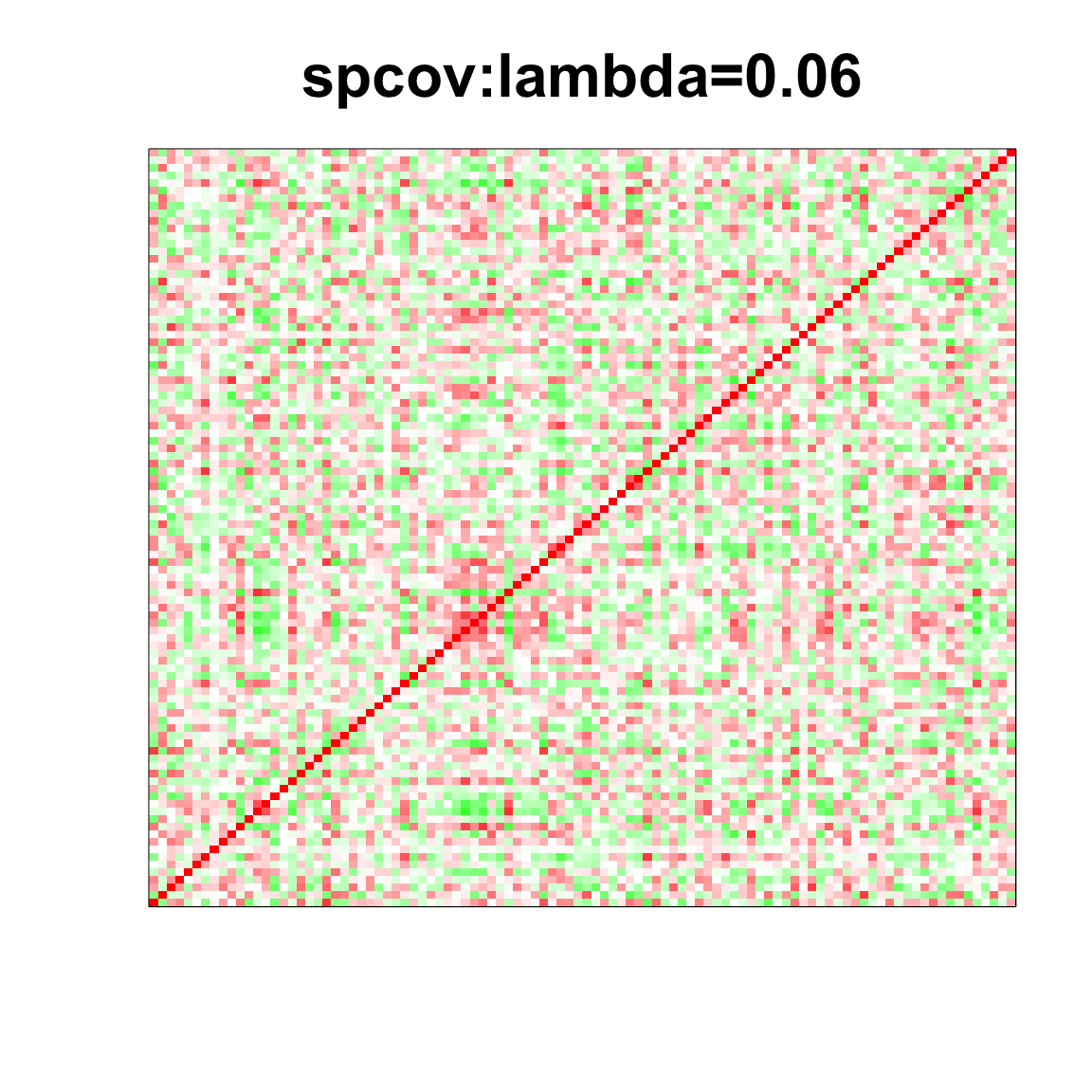
lam <- 0.06
step.size <- 100
tol <- 1e-4
covmat <- cov(data)
mm <- spcov(Sigma=covmat+0.1*diag(1,P), S=covmat+0.1*diag(1,P), lambda=lam * Pat,step.size=step.size, n.inner.steps=200, thr.inner=0, tol.outer=tol, trace=1)
## ---
## using Nesterov, backtracking line search
## ---
## objective: 58.62331
## step size given to GGDescent/Nesterov: 100
## objective: -41.67401 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 20
## step size given to GGDescent/Nesterov: 20
## objective: -42.08276 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 4
## step size given to GGDescent/Nesterov: 4
## objective: -42.09079 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.8
## step size given to GGDescent/Nesterov: 0.8
## objective: -42.09211 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.16
## step size given to GGDescent/Nesterov: 0.16
## objective: -42.09245 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.032
## step size given to GGDescent/Nesterov: 0.032
## objective: -42.09255 ( 200 iterations, max step size: 0.00128 )
## MM converged in 6 steps!
spcor_mat <- cov2cor(mm$Sigma)
#devtools::install_github("kkdey/CorShrink")
#library(CorShrink)
#sessionInfo()
cov_mat <- cov(data);
system.time(strimmer_sample <- corpcor::cov.shrink(data))
## Estimating optimal shrinkage intensity lambda.var (variance vector): 0.6657
##
## Estimating optimal shrinkage intensity lambda (correlation matrix): 0.8459
## user system elapsed
## 0.018 0.000 0.019
system.time(glasso_sample_005 <- glasso::glasso(cov_mat, rho = 0.05))
## user system elapsed
## 0.346 0.001 0.349
system.time(glasso_sample_05 <- glasso::glasso(cov_mat, rho = 0.5))
## user system elapsed
## 0.062 0.000 0.063
system.time(glasso_sample_1 <- glasso::glasso(cov_mat, rho = 1))
## user system elapsed
## 0.002 0.000 0.002
system.time(glasso_sample_10 <- glasso::glasso(cov_mat, rho = 10))
## user system elapsed
## 0.001 0.000 0.002
system.time(cov_sample_ML <- CorShrinkMatrix(cov2cor(cov_mat), nsamp = matrix(n, P, P), ash.control = list(mixcompdist = "normal", nullweight = 10)))
## ash cor only and ash cor PD matrices are different
## user system elapsed
## 2.895 0.028 2.937
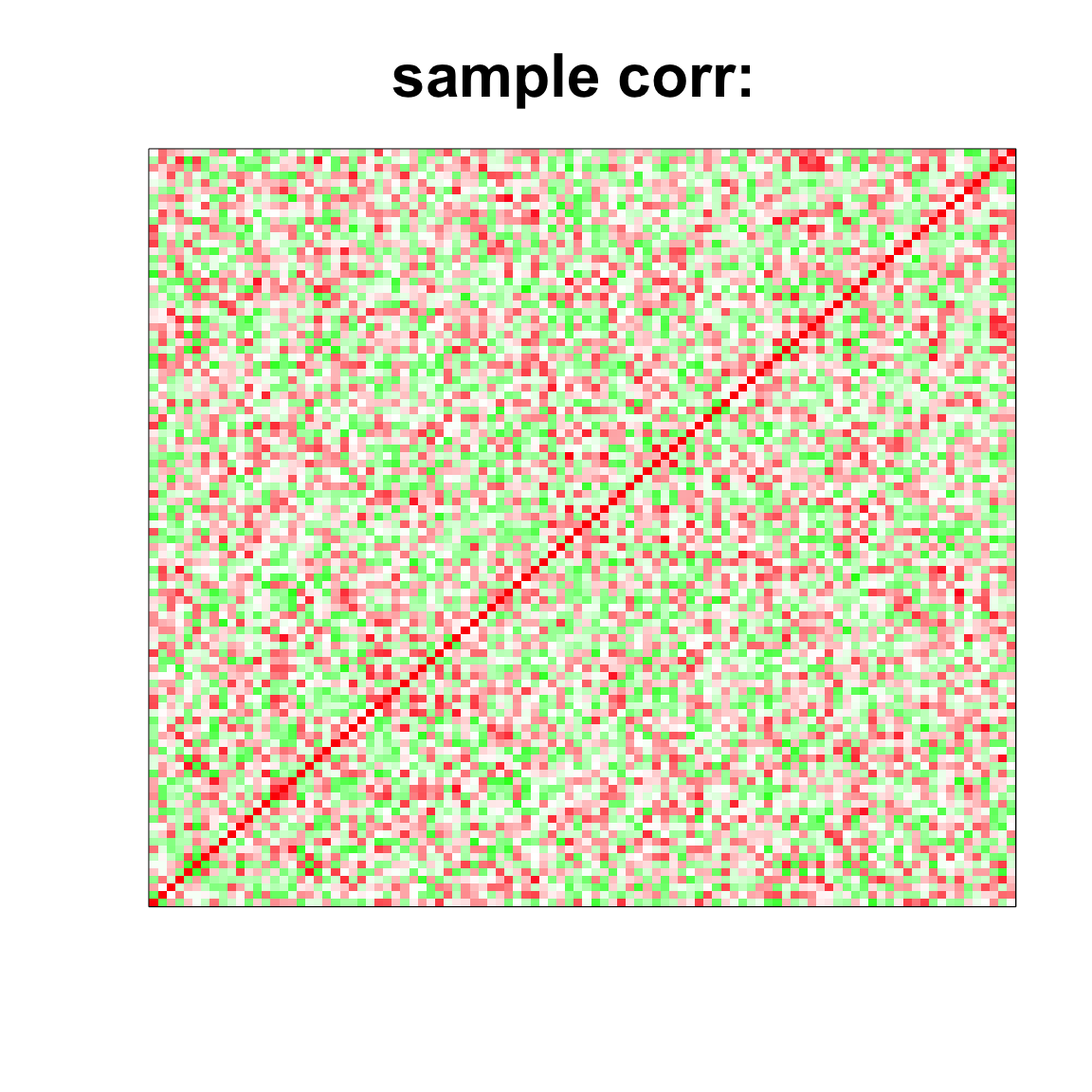
par(mfrow=c(2,2))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
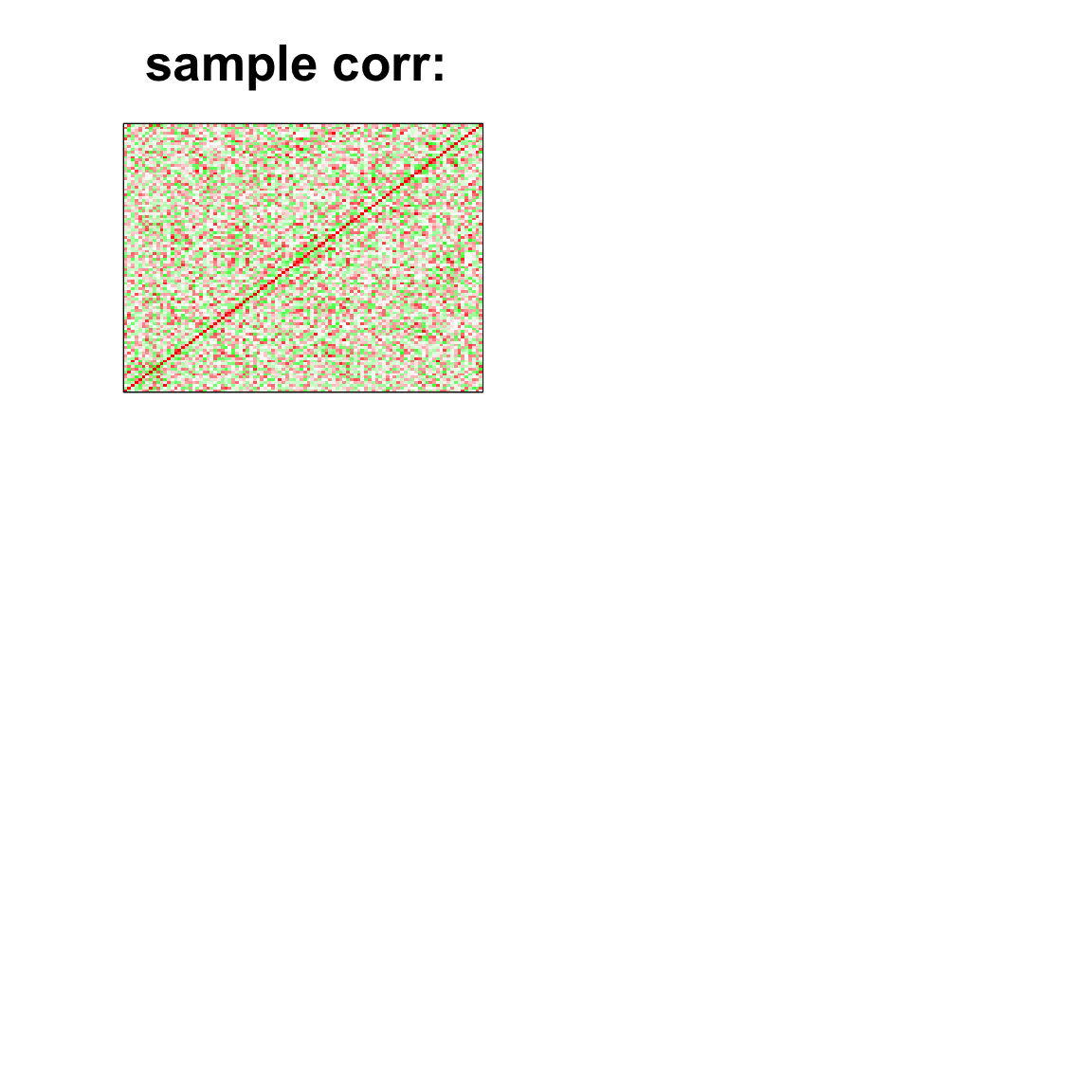
image(as.matrix(cov2cor(cov_mat)),
col=col, main=paste0("sample corr: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
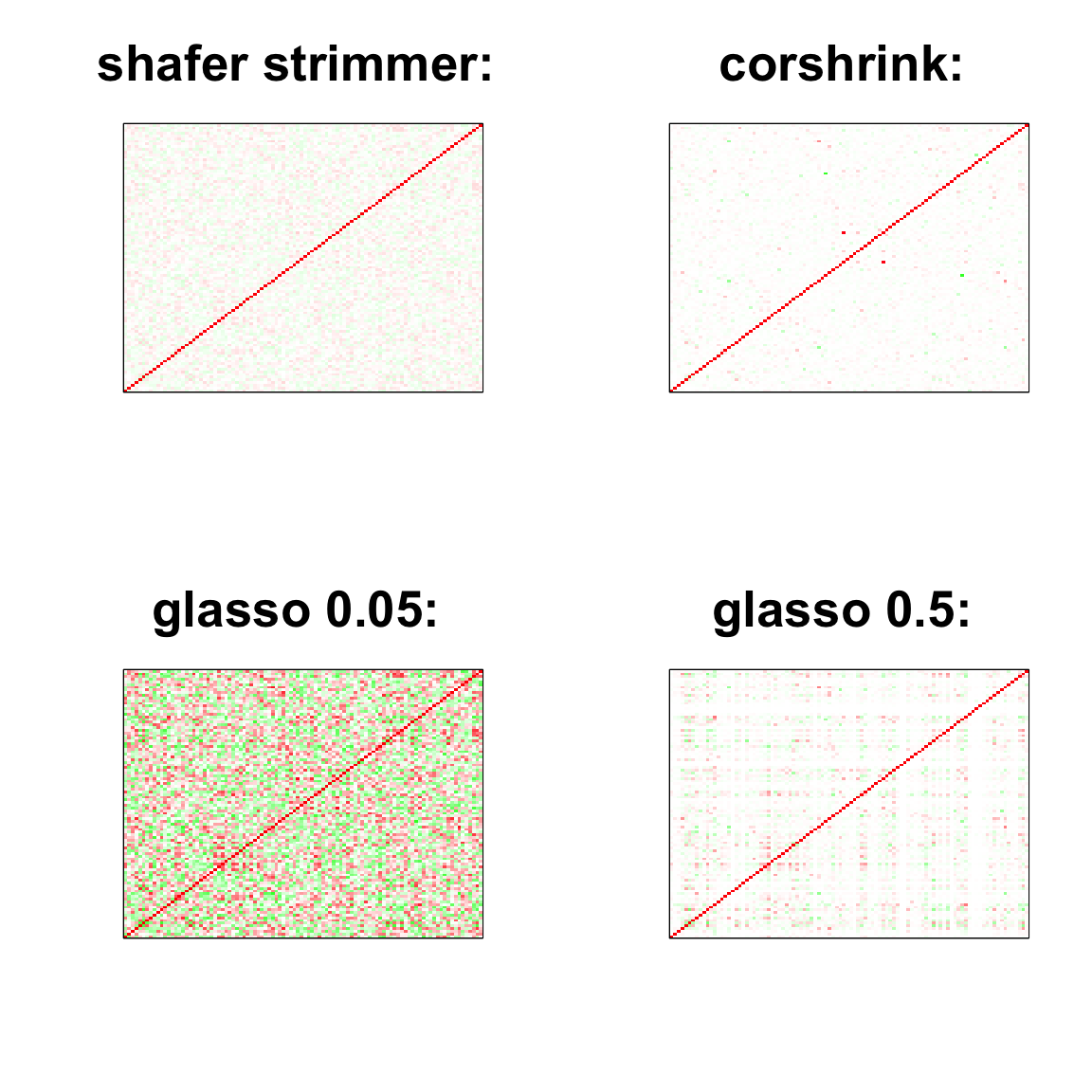
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
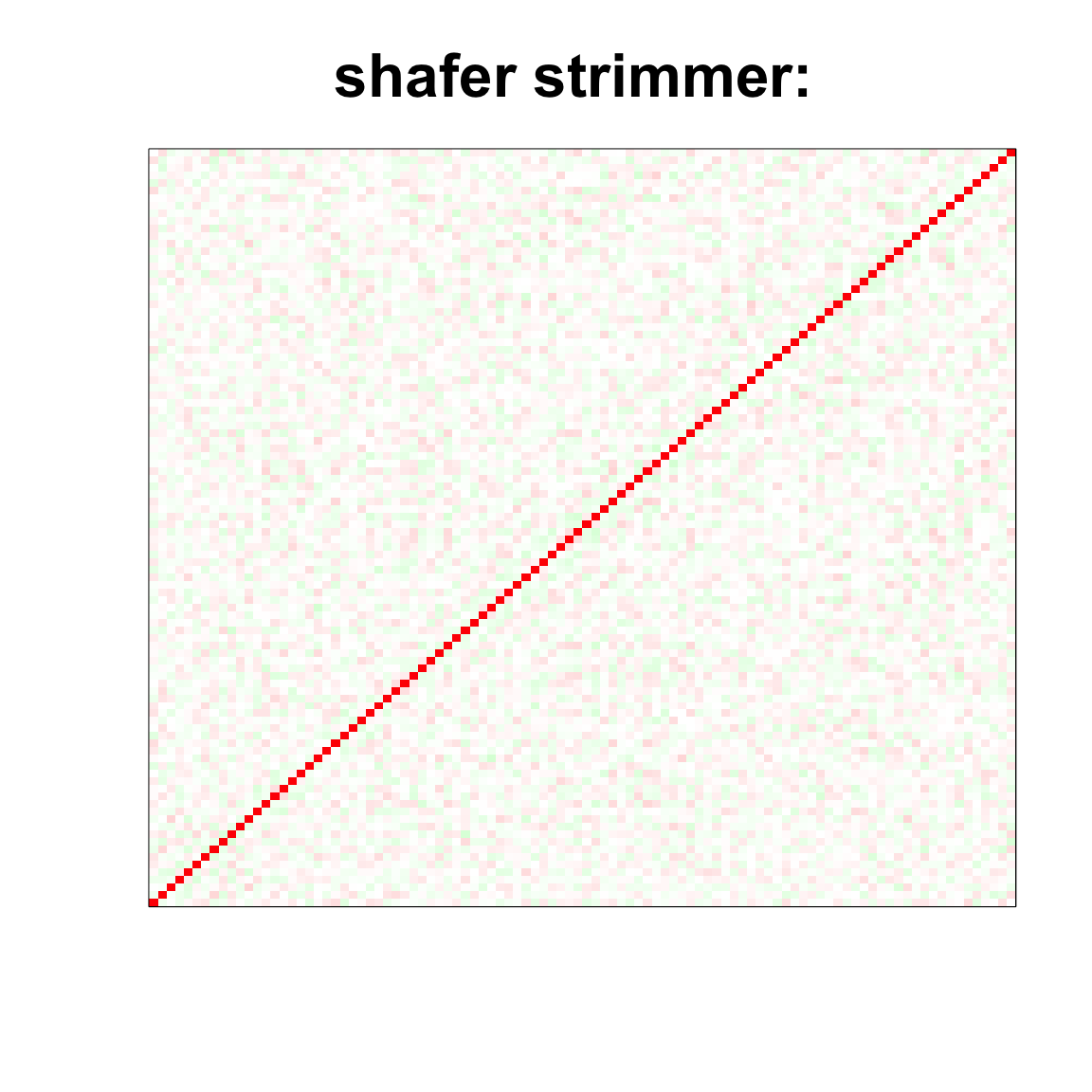
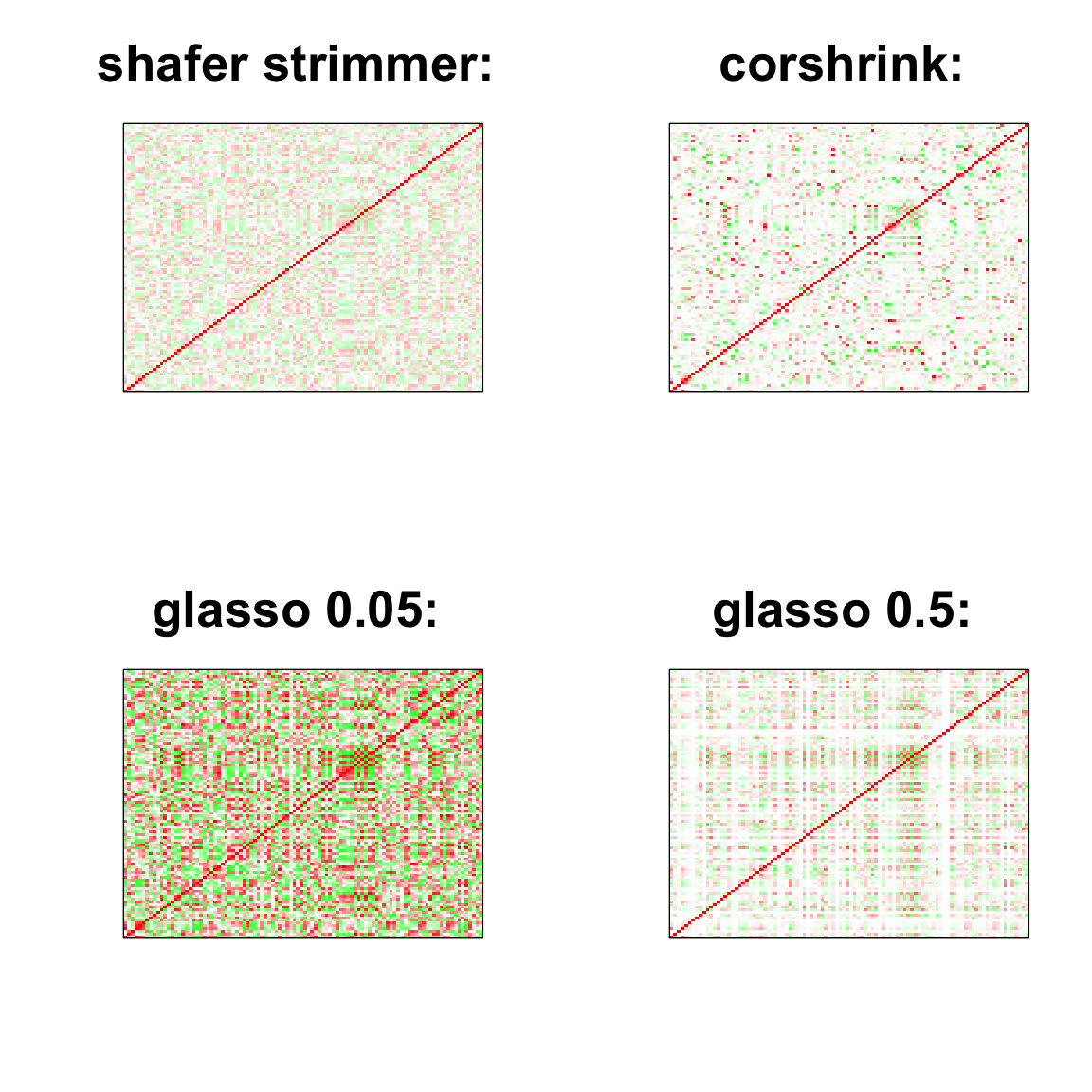
image(as.matrix(cov2cor(strimmer_sample)),
col=col, main=paste0("shafer strimmer: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
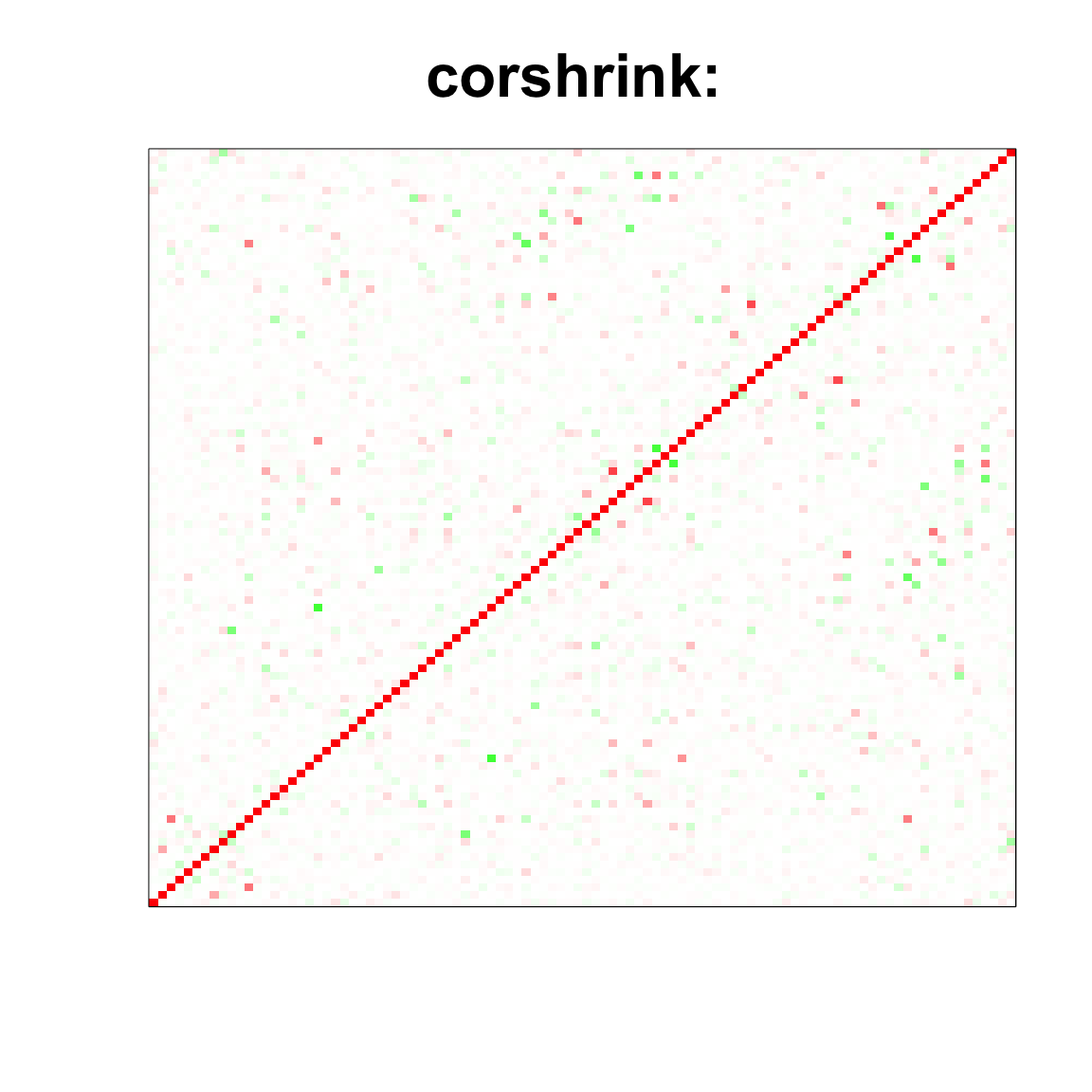
image(as.matrix(cov_sample_ML$ash_cor_only),
col=col, main=paste0("corshrink: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
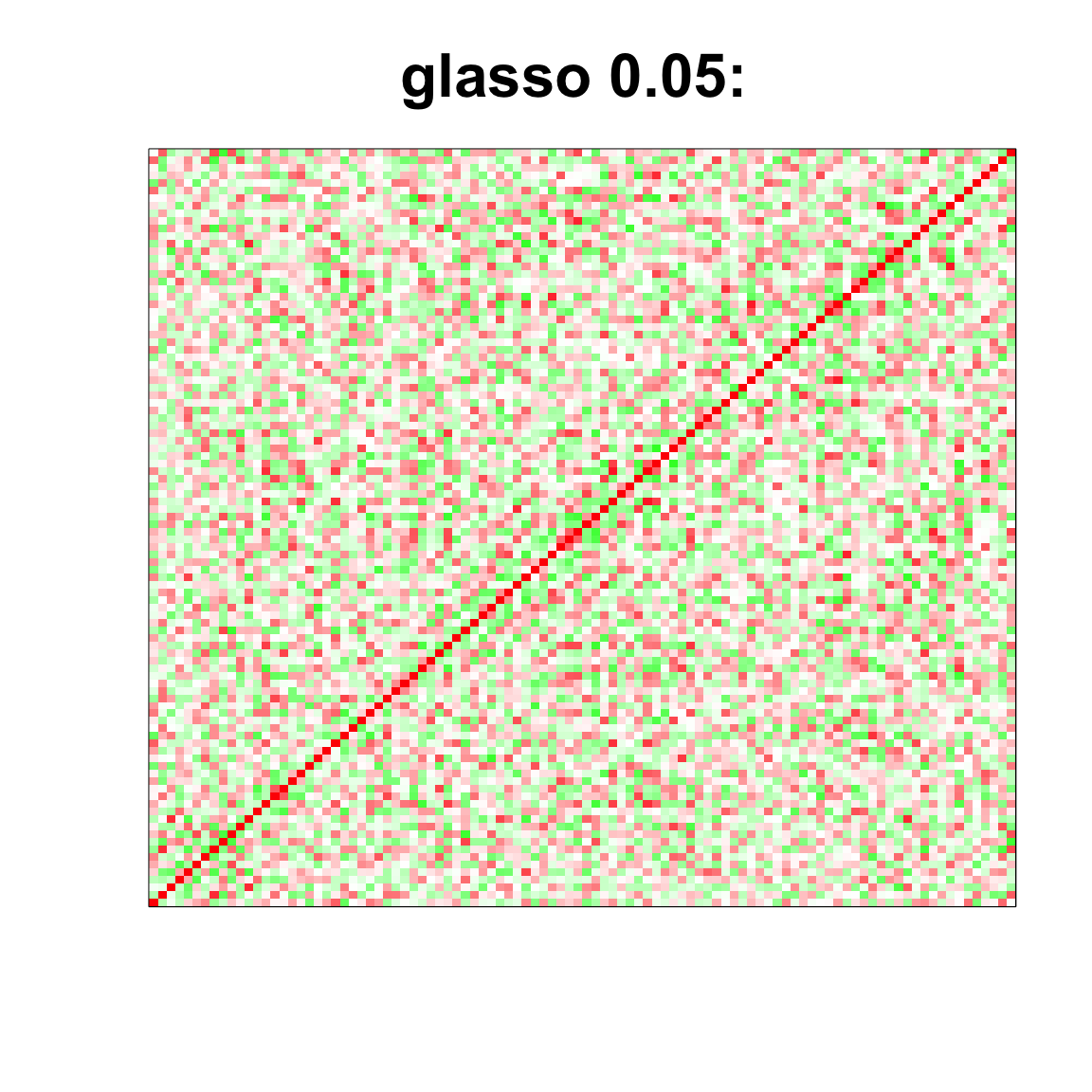
image(as.matrix(cov2cor(glasso_sample_005$w)),
col=col, main=paste0("glasso 0.05: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
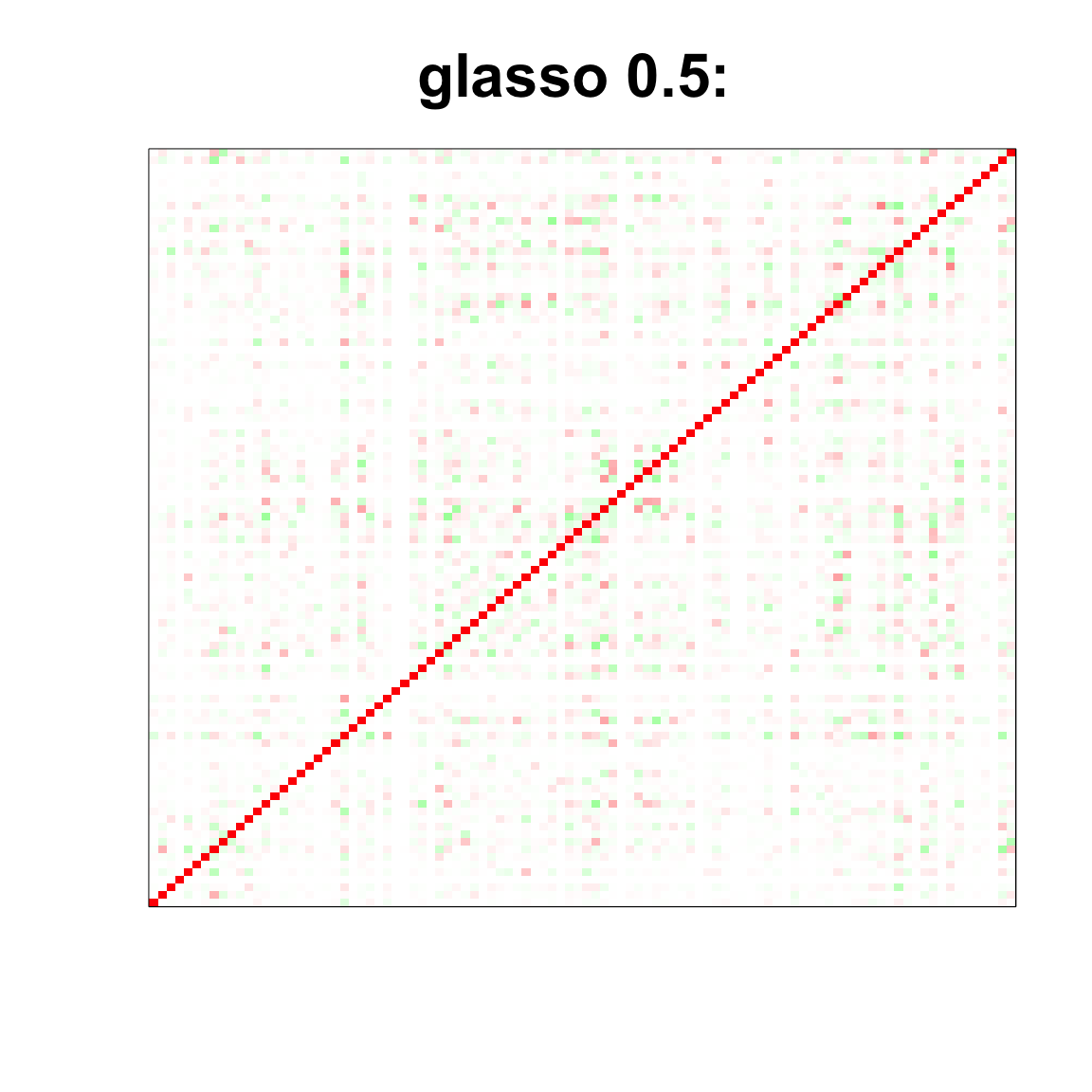
image(as.matrix(cov2cor(glasso_sample_05$w)),
col=col, main=paste0("glasso 0.5: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
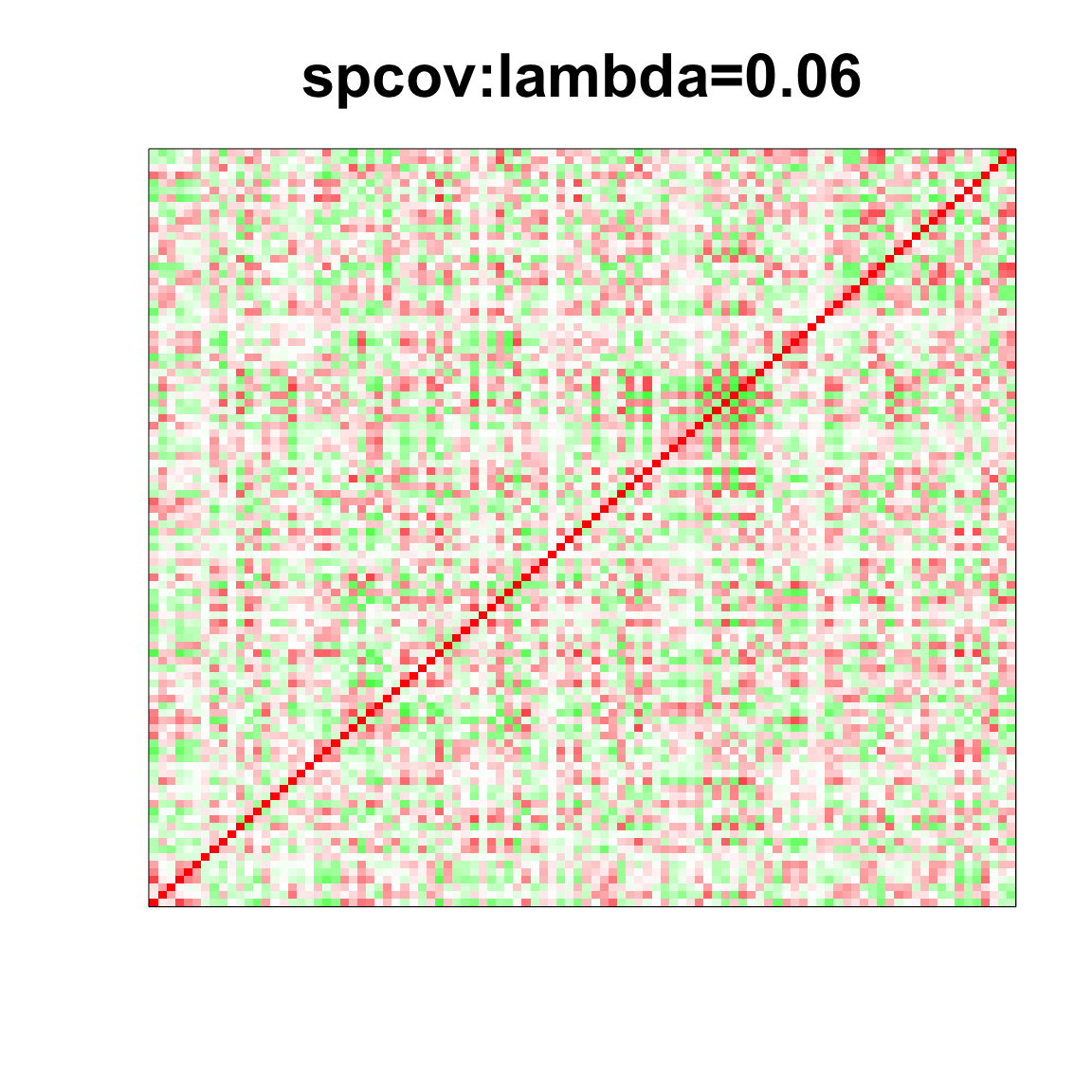
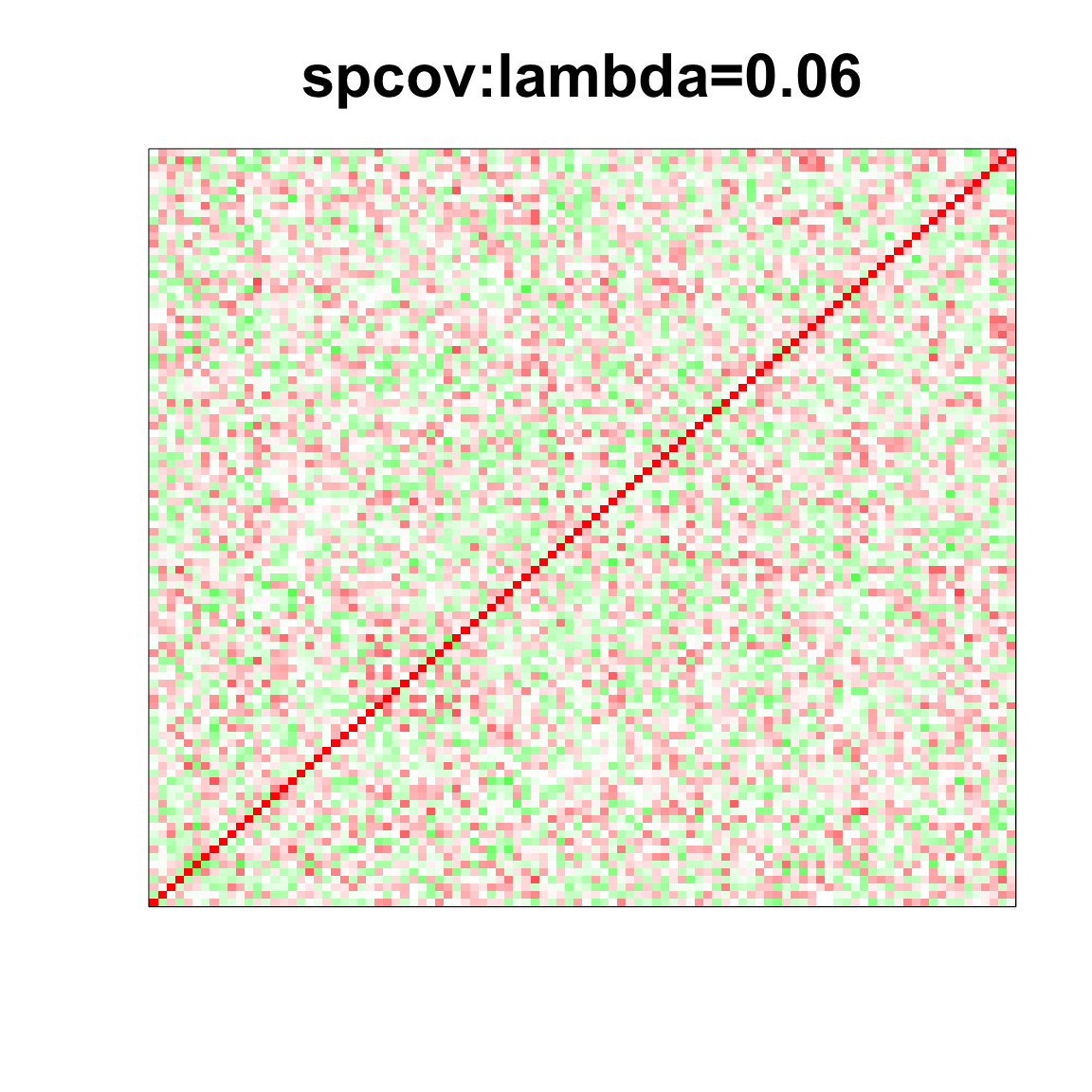
image(as.matrix(spcor_mat),
col=col, main=paste0("spcov:", expression(lambda), "=", lam), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
n = 50, P=100
n <- 50
P <- 100
ll <- DM_graph(n,P,g_type = "band",para = 3)
## Generating data from the multivariate normal distribution with the band graph structure....
## done.
data <- rbind(ll$Xtrain, ll$Xtest)
Sigma <- ll$Sigma
corSigma <- cov2cor(Sigma)
Pat <- matrix(1, P, P)
diag(Pat) <- 0
lam <- 0.06
step.size <- 100
tol <- 1e-4
covmat <- cov(data)
mm <- spcov(Sigma=covmat+0.1*diag(1,P), S=covmat+0.1*diag(1,P), lambda=lam * Pat,step.size=step.size, n.inner.steps=200, thr.inner=0, tol.outer=tol, trace=1)
## ---
## using Nesterov, backtracking line search
## ---
## objective: 77.42316
## step size given to GGDescent/Nesterov: 100
## objective: 42.03639 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 20
## step size given to GGDescent/Nesterov: 20
## objective: 41.16218 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 4
## step size given to GGDescent/Nesterov: 4
## objective: 41.10249 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 0.8
## step size given to GGDescent/Nesterov: 0.8
## objective: 41.09489 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 0.16
## step size given to GGDescent/Nesterov: 0.16
## objective: 41.09326 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 0.032
## step size given to GGDescent/Nesterov: 0.032
## objective: 41.0928 ( 200 iterations, max step size: 0.0064 )
## step size given to GGDescent/Nesterov: 0.032
## objective: 41.09266 ( 200 iterations, max step size: 0.0064 )
## step size given to GGDescent/Nesterov: 0.032
## objective: 41.09262 ( 200 iterations, max step size: 0.0064 )
## MM converged in 8 steps!
spcor_mat <- cov2cor(mm$Sigma)
#devtools::install_github("kkdey/CorShrink")
#library(CorShrink)
#sessionInfo()
cov_mat <- cov(data);
system.time(strimmer_sample <- corpcor::cov.shrink(data))
## Estimating optimal shrinkage intensity lambda.var (variance vector): 1
##
## Estimating optimal shrinkage intensity lambda (correlation matrix): 0.7994
## user system elapsed
## 0.079 0.000 0.080
system.time(glasso_sample_005 <- glasso::glasso(cov_mat, rho = 0.05))
## user system elapsed
## 0.154 0.000 0.155
system.time(glasso_sample_05 <- glasso::glasso(cov_mat, rho = 0.5))
## user system elapsed
## 0.002 0.000 0.002
system.time(glasso_sample_1 <- glasso::glasso(cov_mat, rho = 1))
## user system elapsed
## 0.001 0.000 0.002
system.time(glasso_sample_10 <- glasso::glasso(cov_mat, rho = 10))
## user system elapsed
## 0.001 0.000 0.002
system.time(cov_sample_ML <- CorShrinkMatrix(cov2cor(cov_mat), nsamp = matrix(n, P, P), ash.control = list(mixcompdist = "normal", nullweight = 10)))
## ash cor only and ash cor PD matrices are different
## user system elapsed
## 3.506 0.072 3.590
par(mfrow=c(2,2))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(cov_mat)),
col=col, main=paste0("sample corr: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(strimmer_sample)),
col=col, main=paste0("shafer strimmer: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
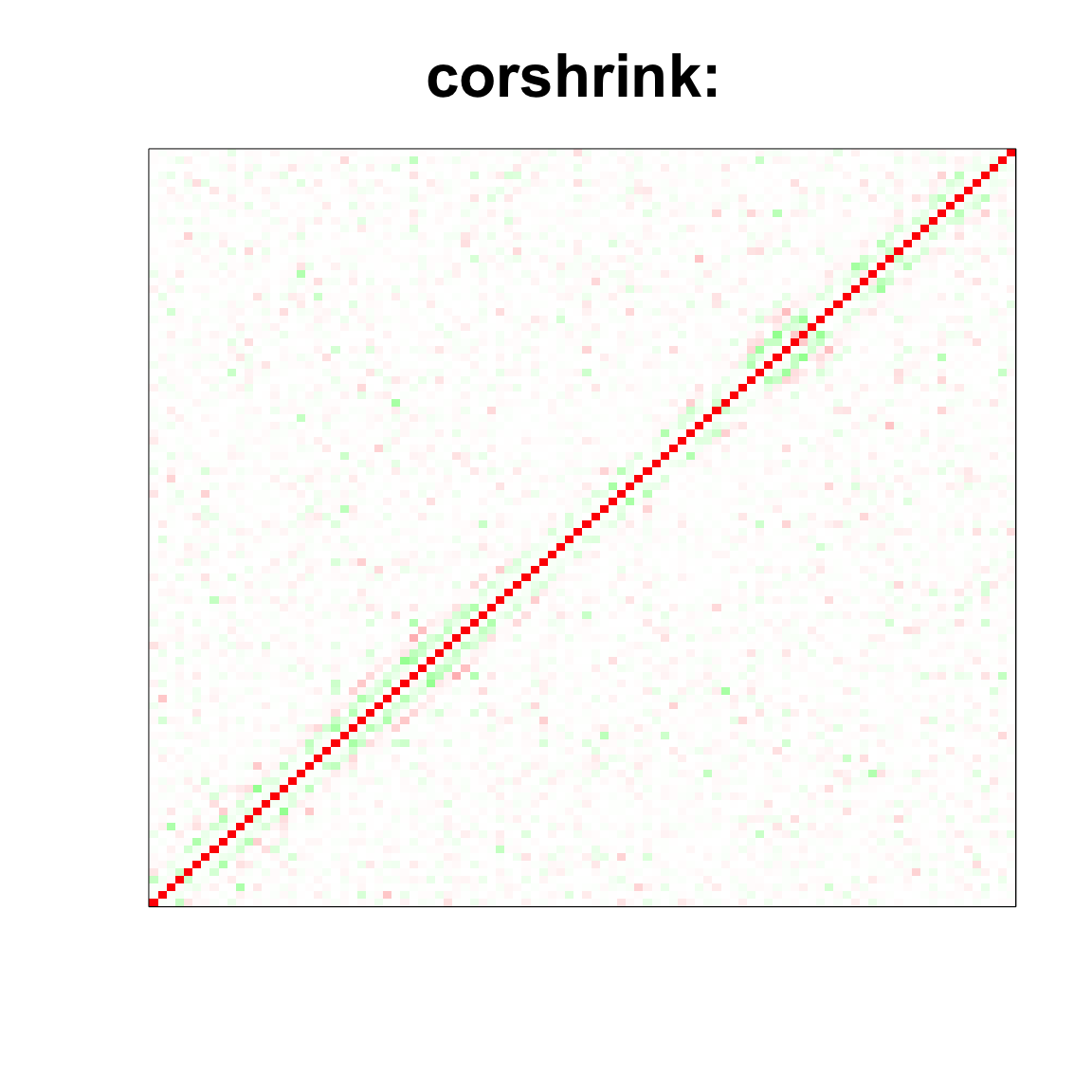
image(as.matrix(cov_sample_ML$ash_cor_only),
col=col, main=paste0("corshrink: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
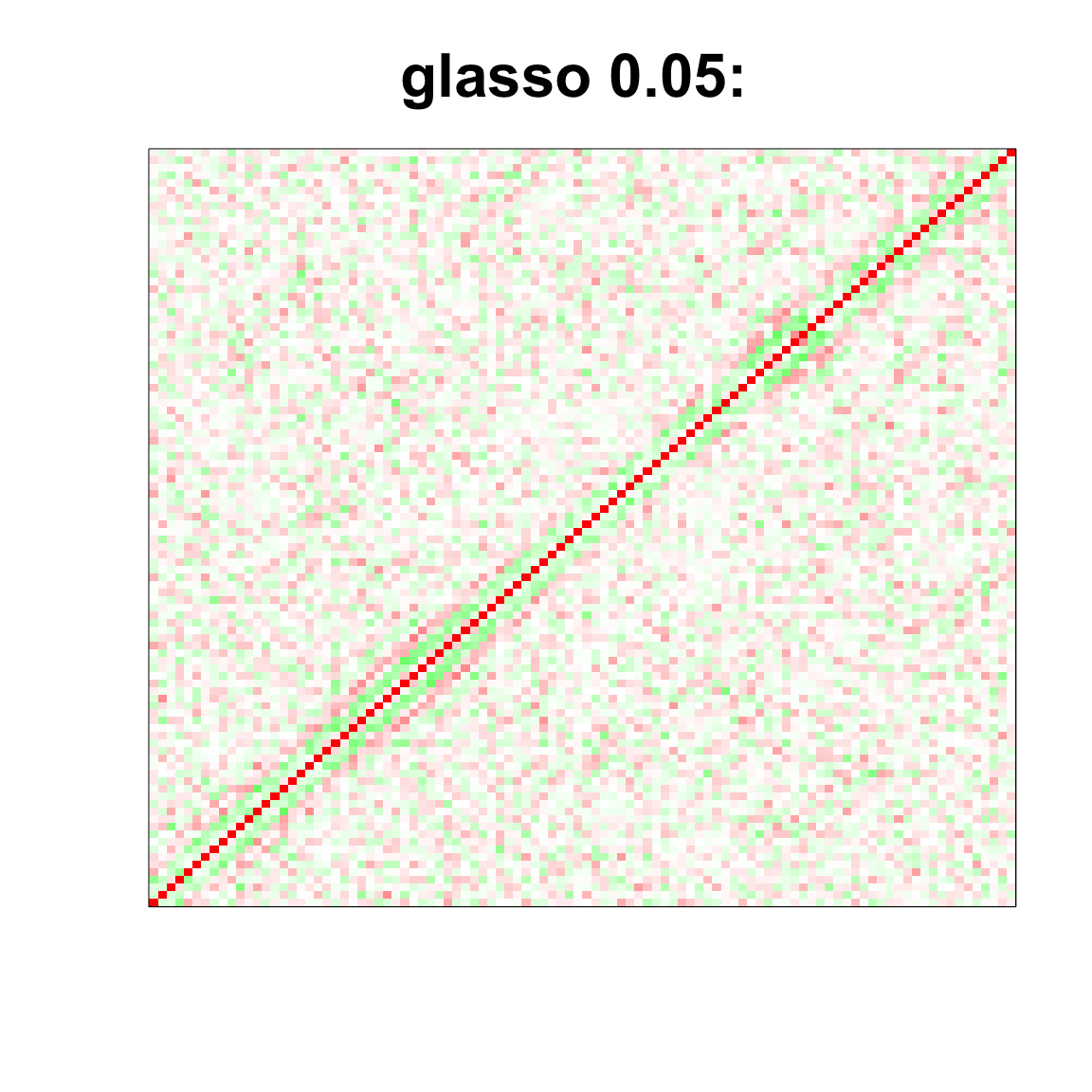
image(as.matrix(cov2cor(glasso_sample_005$w)),
col=col, main=paste0("glasso 0.05: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
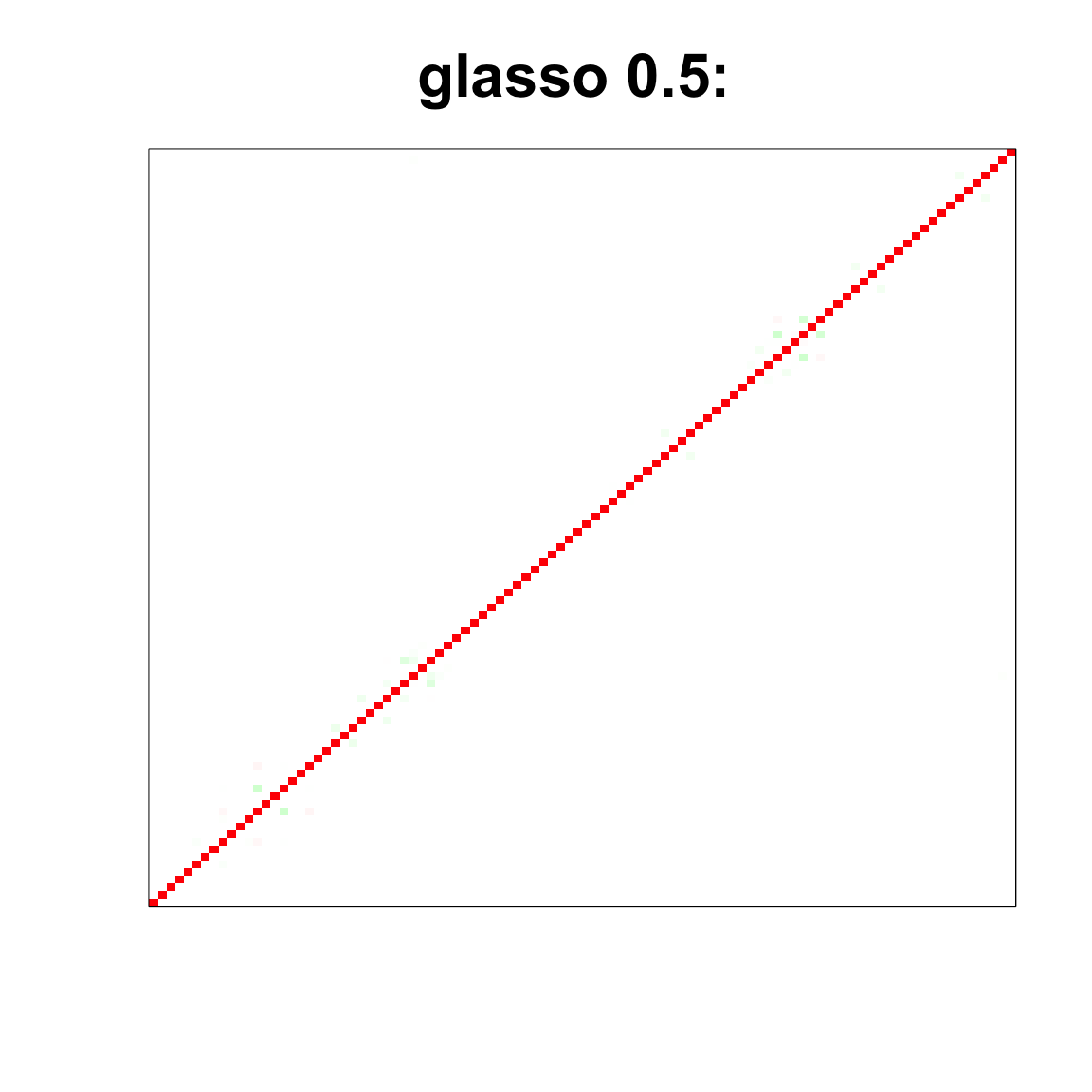
image(as.matrix(cov2cor(glasso_sample_05$w)),
col=col, main=paste0("glasso 0.5: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
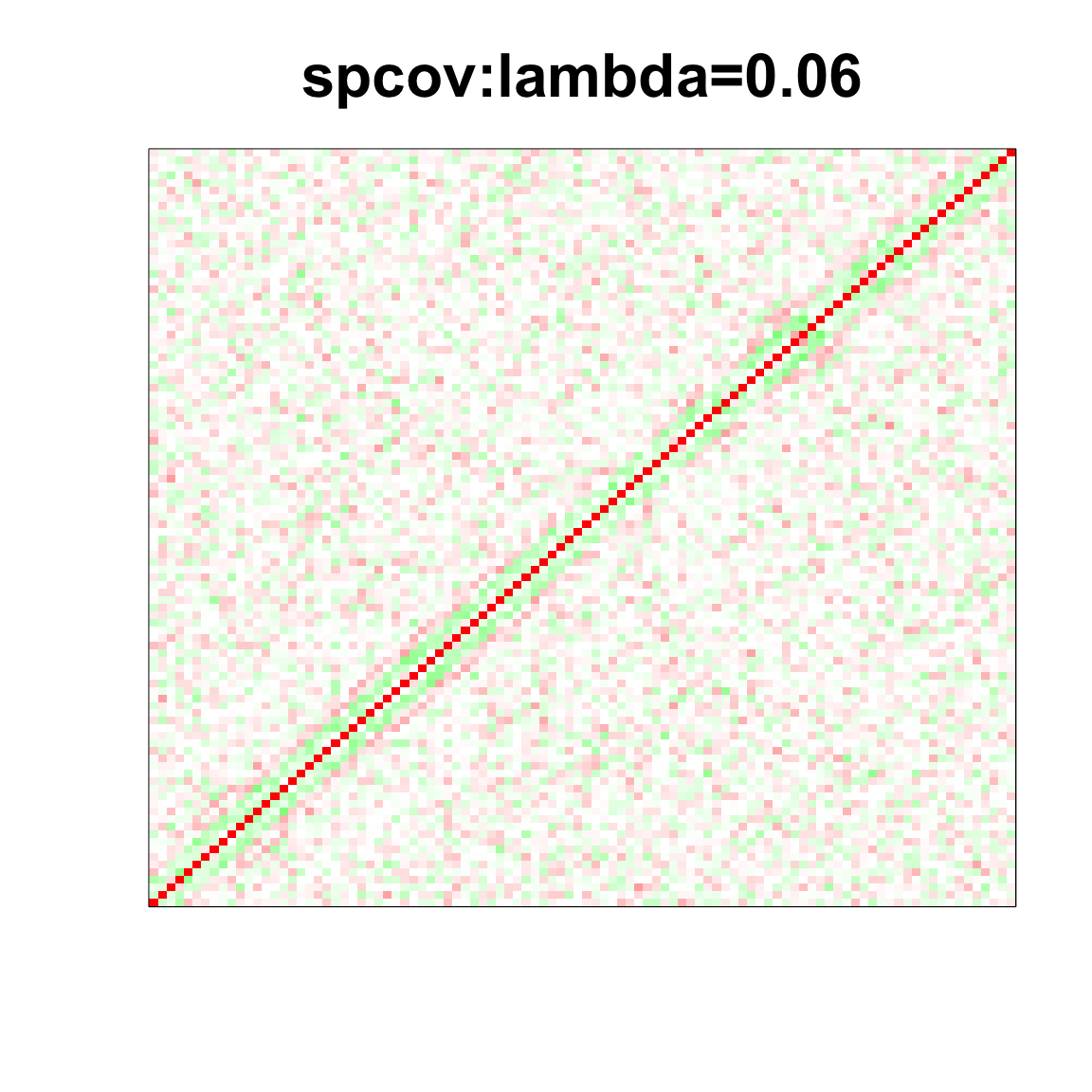
image(as.matrix(spcor_mat),
col=col, main=paste0("spcov:", expression(lambda), "=", lam), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
n = 5, P=100
n <- 5
P <- 100
ll <- DM_graph(n,P,g_type = "band",para = 3)
## Generating data from the multivariate normal distribution with the band graph structure....
## done.
data <- rbind(ll$Xtrain, ll$Xtest)
Sigma <- ll$Sigma
corSigma <- cov2cor(Sigma)
Pat <- matrix(1, P, P)
diag(Pat) <- 0
lam <- 0.06
step.size <- 100
tol <- 1e-4
covmat <- cov(data)
mm <- spcov(Sigma=covmat+0.1*diag(1,P), S=covmat+0.1*diag(1,P), lambda=lam * Pat,step.size=step.size, n.inner.steps=200, thr.inner=0, tol.outer=tol, trace=1)
## ---
## using Nesterov, backtracking line search
## ---
## objective: 118.8094
## step size given to GGDescent/Nesterov: 100
## objective: -60.78012 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 20
## step size given to GGDescent/Nesterov: 20
## objective: -63.18863 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 4
## step size given to GGDescent/Nesterov: 4
## objective: -63.19932 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.8
## step size given to GGDescent/Nesterov: 0.8
## objective: -63.20103 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.16
## step size given to GGDescent/Nesterov: 0.16
## objective: -63.20148 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.032
## step size given to GGDescent/Nesterov: 0.032
## objective: -63.20161 ( 200 iterations, max step size: 0.00128 )
## step size given to GGDescent/Nesterov: 0.032
## objective: -63.20164 ( 200 iterations, max step size: 0.00128 )
## MM converged in 7 steps!
spcor_mat <- cov2cor(mm$Sigma)
#devtools::install_github("kkdey/CorShrink")
#library(CorShrink)
#sessionInfo()
cov_mat <- cov(data);
system.time(strimmer_sample <- corpcor::cov.shrink(data))
## Estimating optimal shrinkage intensity lambda.var (variance vector): 0.7966
##
## Estimating optimal shrinkage intensity lambda (correlation matrix): 0.7162
## user system elapsed
## 0.009 0.000 0.009
system.time(glasso_sample_005 <- glasso::glasso(cov_mat, rho = 0.05))
## user system elapsed
## 0.628 0.001 0.631
system.time(glasso_sample_05 <- glasso::glasso(cov_mat, rho = 0.5))
## user system elapsed
## 0.054 0.000 0.054
system.time(glasso_sample_1 <- glasso::glasso(cov_mat, rho = 1))
## user system elapsed
## 0.012 0.000 0.012
system.time(glasso_sample_10 <- glasso::glasso(cov_mat, rho = 10))
## user system elapsed
## 0.001 0.000 0.002
system.time(cov_sample_ML <- CorShrinkMatrix(cov2cor(cov_mat), nsamp = matrix(n, P, P), ash.control = list(mixcompdist = "normal", nullweight = 10)))
## ash cor only and ash cor PD matrices are different
## user system elapsed
## 1.872 0.035 1.915
par(mfrow=c(2,2))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
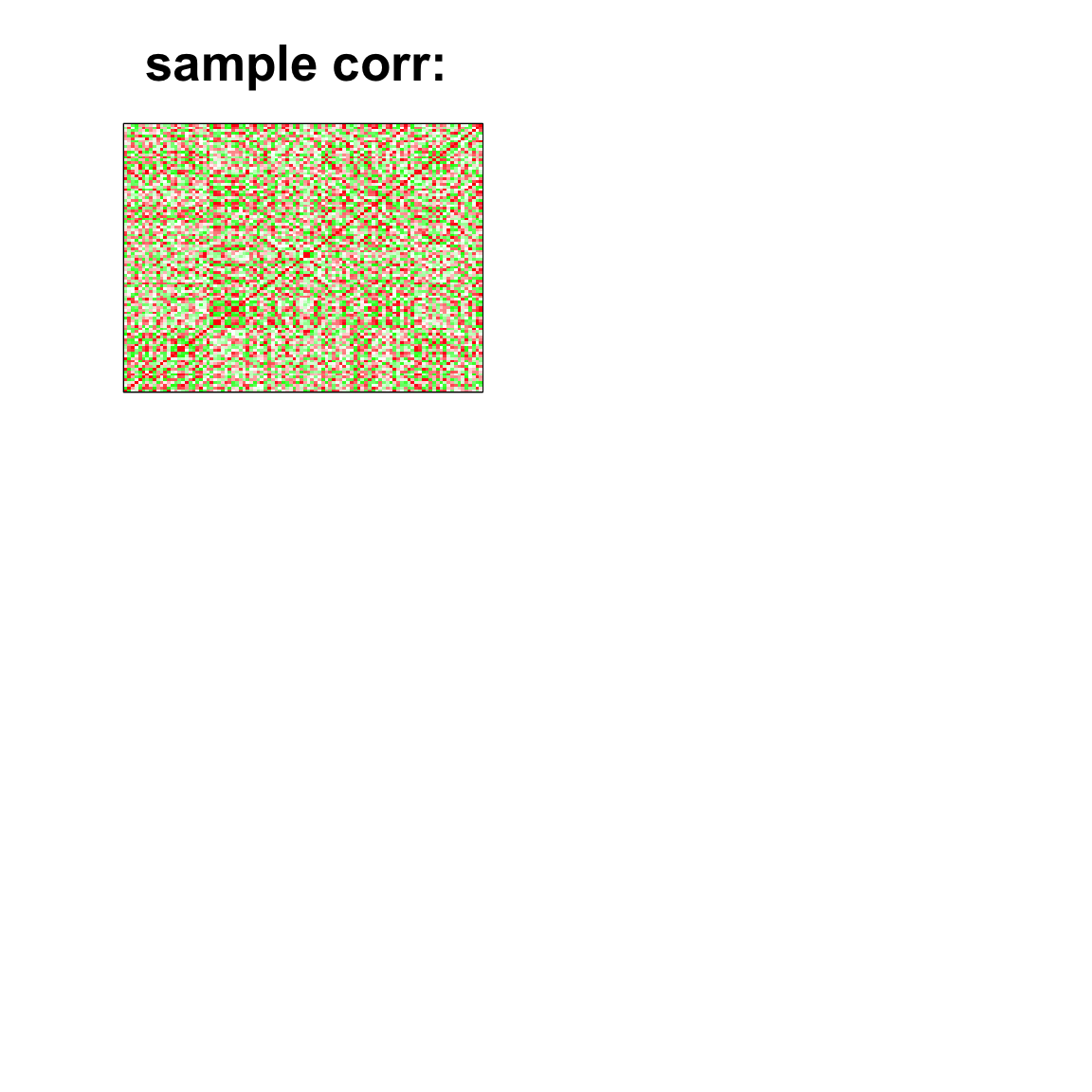
image(as.matrix(cov2cor(cov_mat)),
col=col, main=paste0("sample corr: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
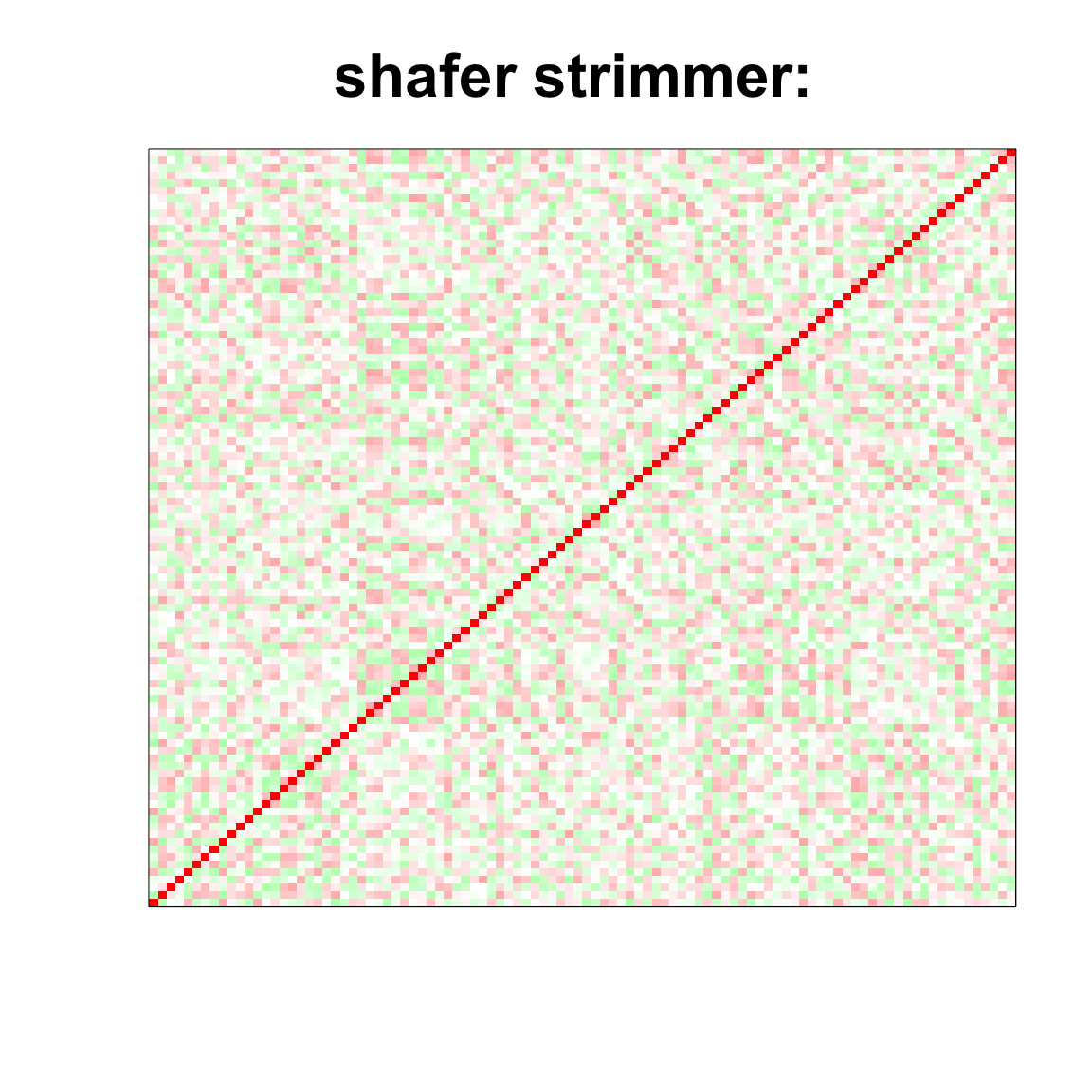
image(as.matrix(cov2cor(strimmer_sample)),
col=col, main=paste0("shafer strimmer: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
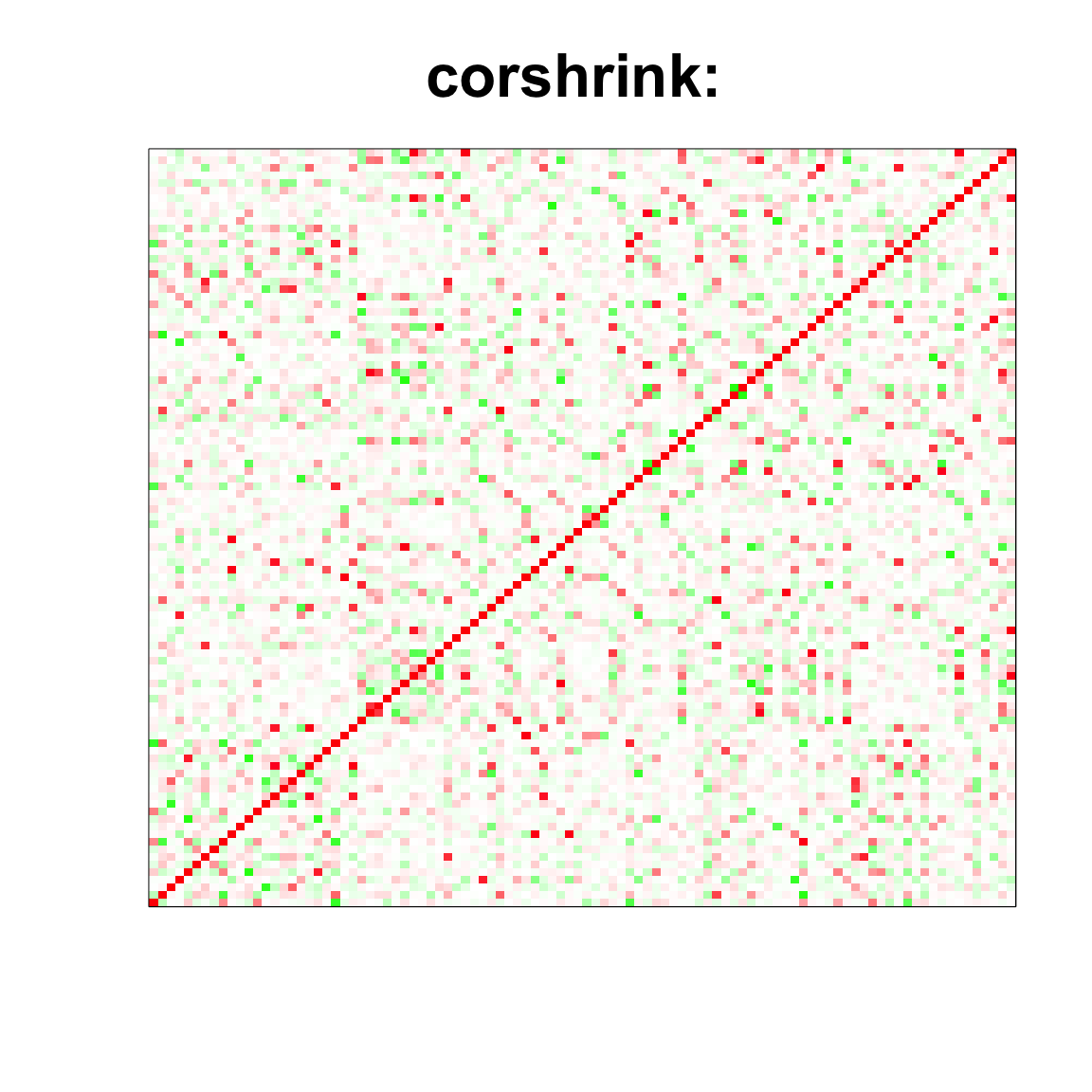
image(as.matrix(cov_sample_ML$ash_cor_only),
col=col, main=paste0("corshrink: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(glasso_sample_005$w)),
col=col, main=paste0("glasso 0.05: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(glasso_sample_05$w)),
col=col, main=paste0("glasso 0.5: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(spcor_mat),
col=col, main=paste0("spcov:", expression(lambda), "=", lam), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
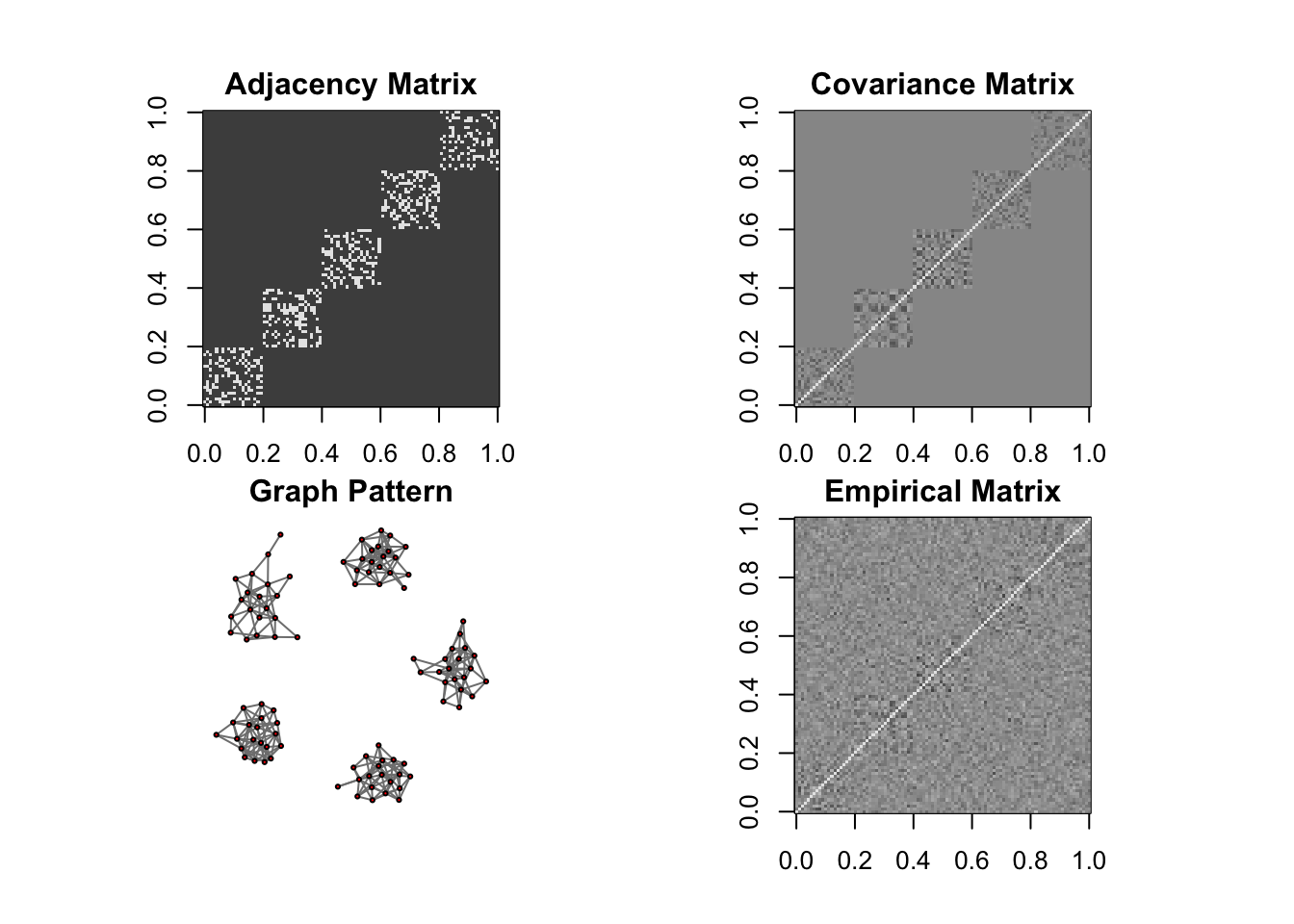
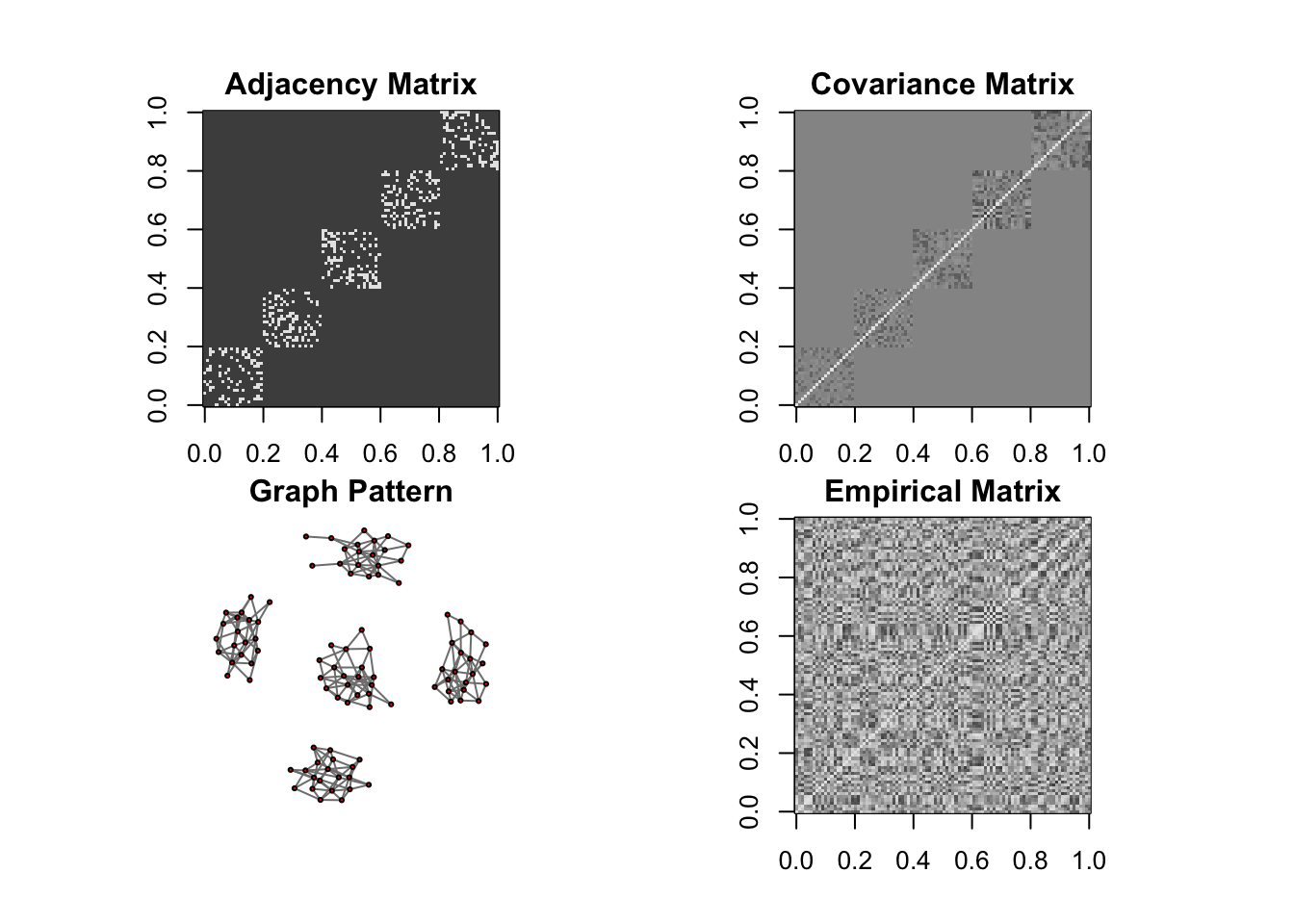
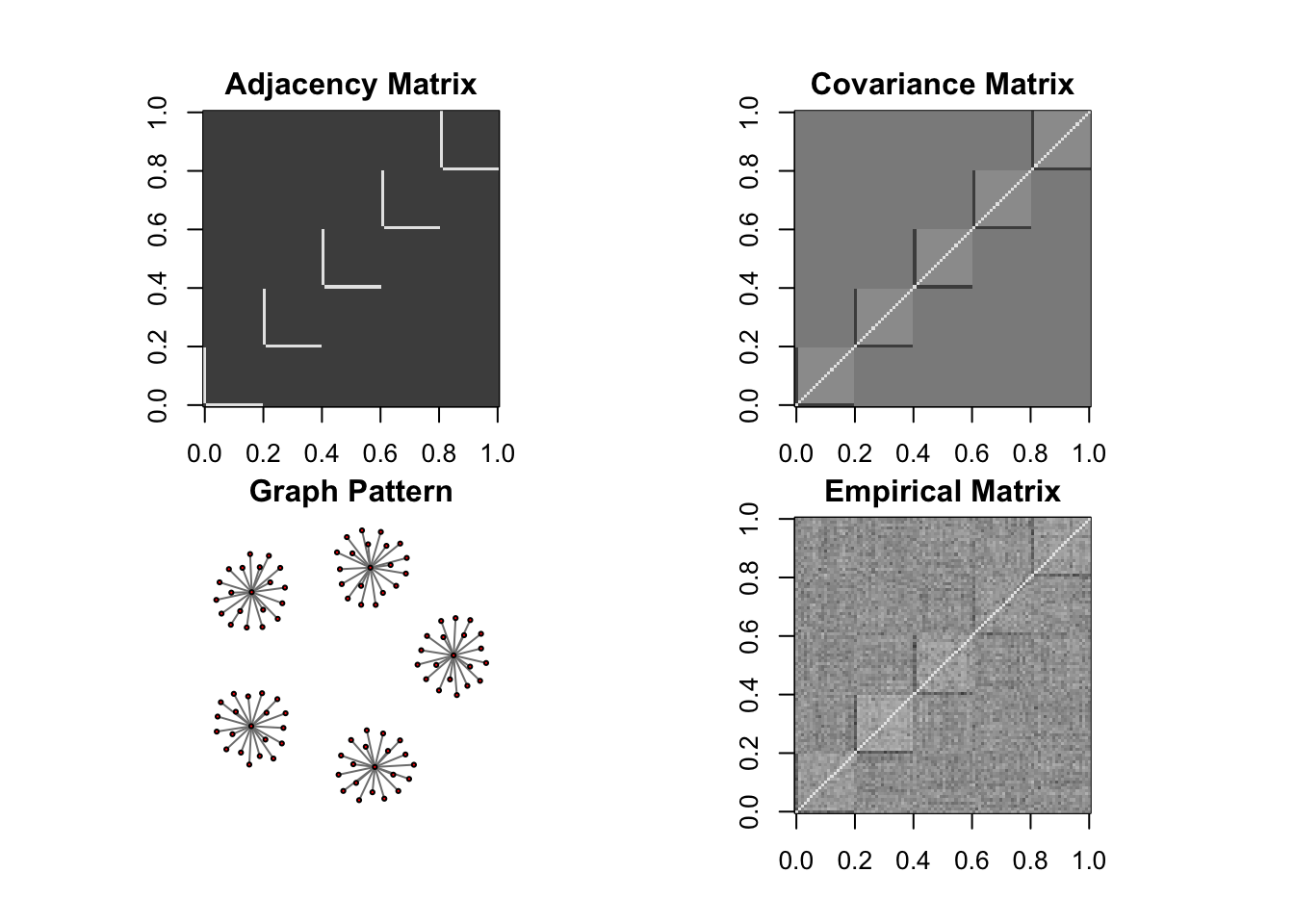
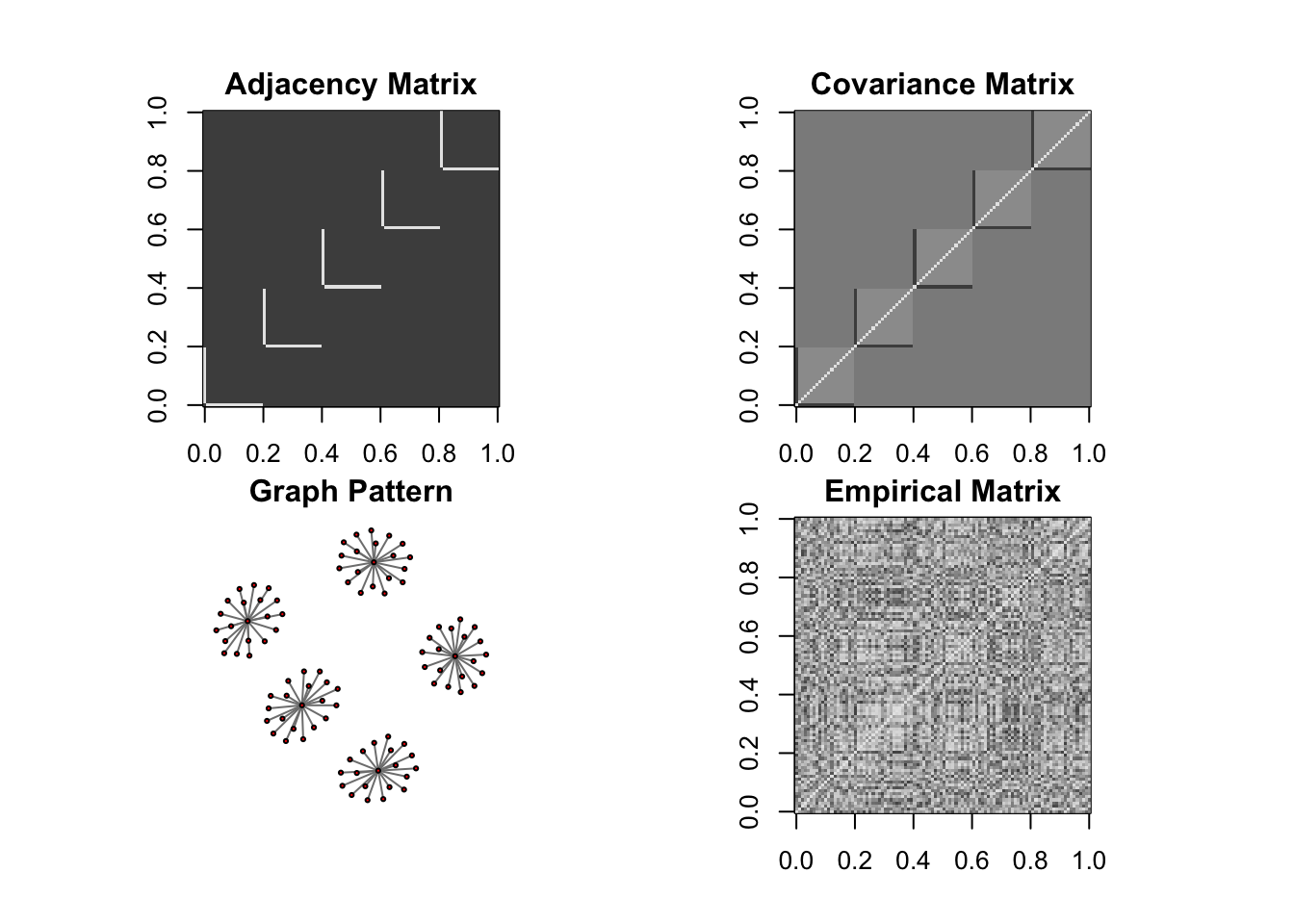
DM graph (cluster)
n = 10, P=100
n <- 10
P <- 100
ll <- DM_graph(n,P,g_type = "cluster", para = 5)
## Generating data from the multivariate normal distribution with the cluster graph structure....
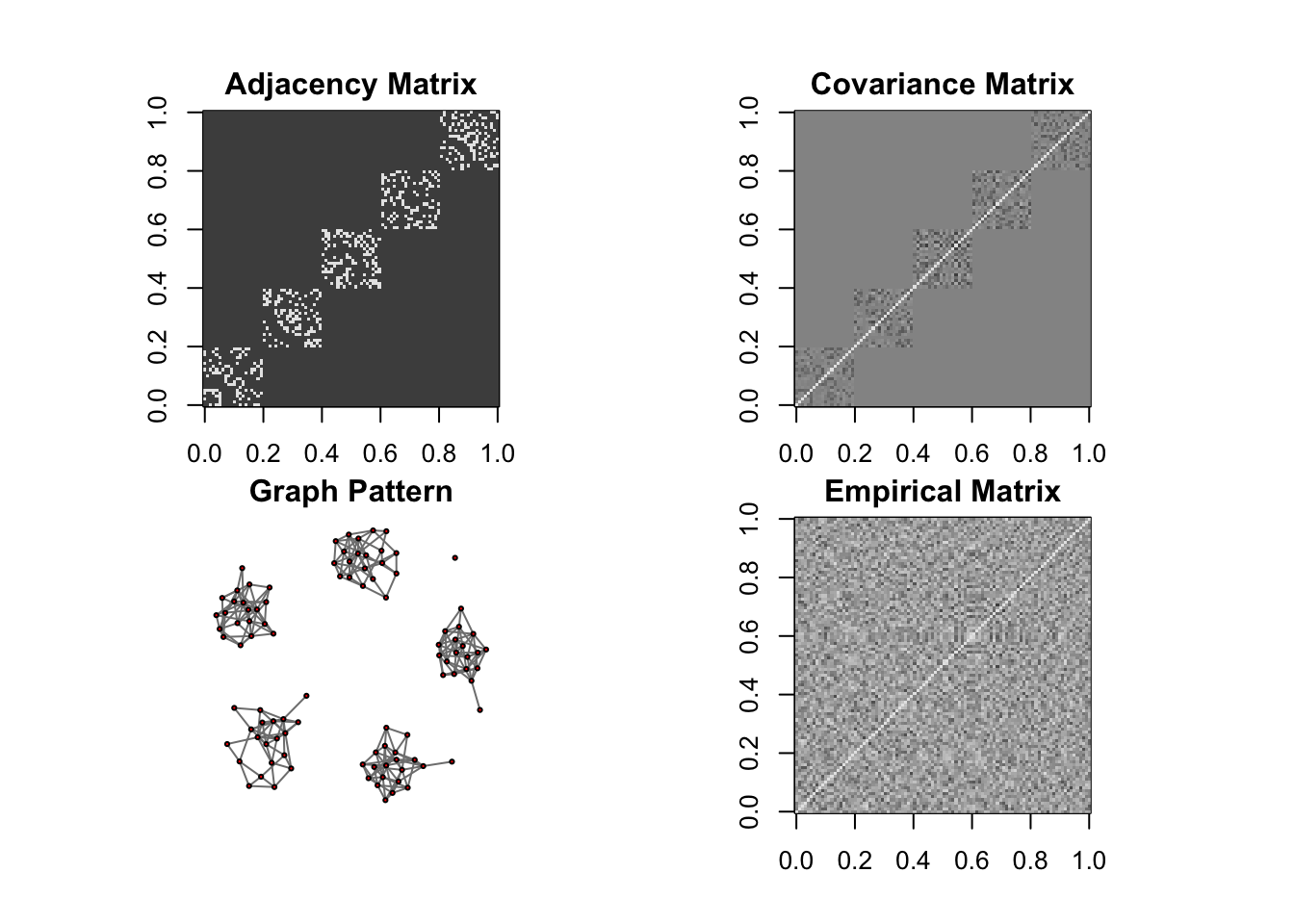
## done.
data <- rbind(ll$Xtrain, ll$Xtest)
Sigma <- ll$Sigma
corSigma <- cov2cor(Sigma)
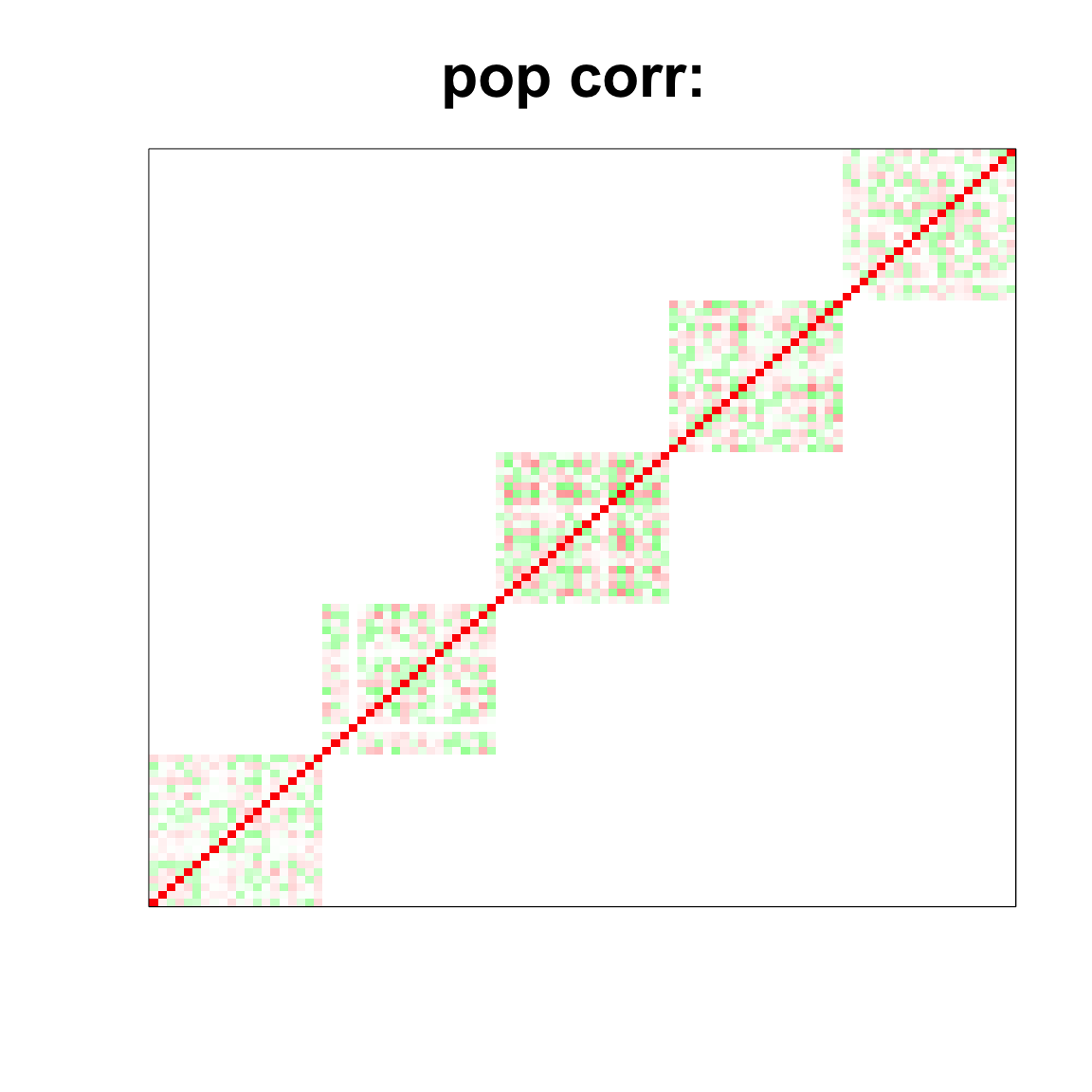
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(corSigma),
col=col, main=paste0("pop corr: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
Pat <- matrix(1, P, P)
diag(Pat) <- 0
lam <- 0.06
step.size <- 100
tol <- 1e-4
covmat <- cov(data)
mm <- spcov(Sigma=covmat+0.1*diag(1,P), S=covmat+0.1*diag(1,P), lambda=lam * Pat,step.size=step.size, n.inner.steps=200, thr.inner=0, tol.outer=tol, trace=1)
## ---
## using Nesterov, backtracking line search
## ---
## objective: 64.48307
## step size given to GGDescent/Nesterov: 100
## objective: -38.88554 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 20
## step size given to GGDescent/Nesterov: 20
## objective: -39.44978 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 4
## step size given to GGDescent/Nesterov: 4
## objective: -39.45928 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.8
## step size given to GGDescent/Nesterov: 0.8
## objective: -39.46073 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.16
## step size given to GGDescent/Nesterov: 0.16
## objective: -39.4611 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.032
## step size given to GGDescent/Nesterov: 0.032
## objective: -39.4612 ( 200 iterations, max step size: 0.00128 )
## step size given to GGDescent/Nesterov: 0.032
## objective: -39.46123 ( 200 iterations, max step size: 0.00128 )
## MM converged in 7 steps!
spcor_mat <- cov2cor(mm$Sigma)
#devtools::install_github("kkdey/CorShrink")
#library(CorShrink)
#sessionInfo()
cov_mat <- cov(data);
system.time(strimmer_sample <- corpcor::cov.shrink(data))
## Estimating optimal shrinkage intensity lambda.var (variance vector): 0.7148
##
## Estimating optimal shrinkage intensity lambda (correlation matrix): 0.8998
## user system elapsed
## 0.002 0.000 0.001
system.time(glasso_sample_005 <- glasso::glasso(cov_mat, rho = 0.05))
## user system elapsed
## 0.378 0.001 0.380
system.time(glasso_sample_05 <- glasso::glasso(cov_mat, rho = 0.5))
## user system elapsed
## 0.032 0.000 0.033
system.time(glasso_sample_1 <- glasso::glasso(cov_mat, rho = 1))
## user system elapsed
## 0.002 0.000 0.002
system.time(glasso_sample_10 <- glasso::glasso(cov_mat, rho = 10))
## user system elapsed
## 0.001 0.000 0.001
system.time(cov_sample_ML <- CorShrinkMatrix(cov2cor(cov_mat), nsamp = matrix(n, P, P), ash.control = list(mixcompdist = "normal", nullweight = 10)))
## ash cor only and ash cor PD matrices are different
## user system elapsed
## 2.200 0.063 2.339
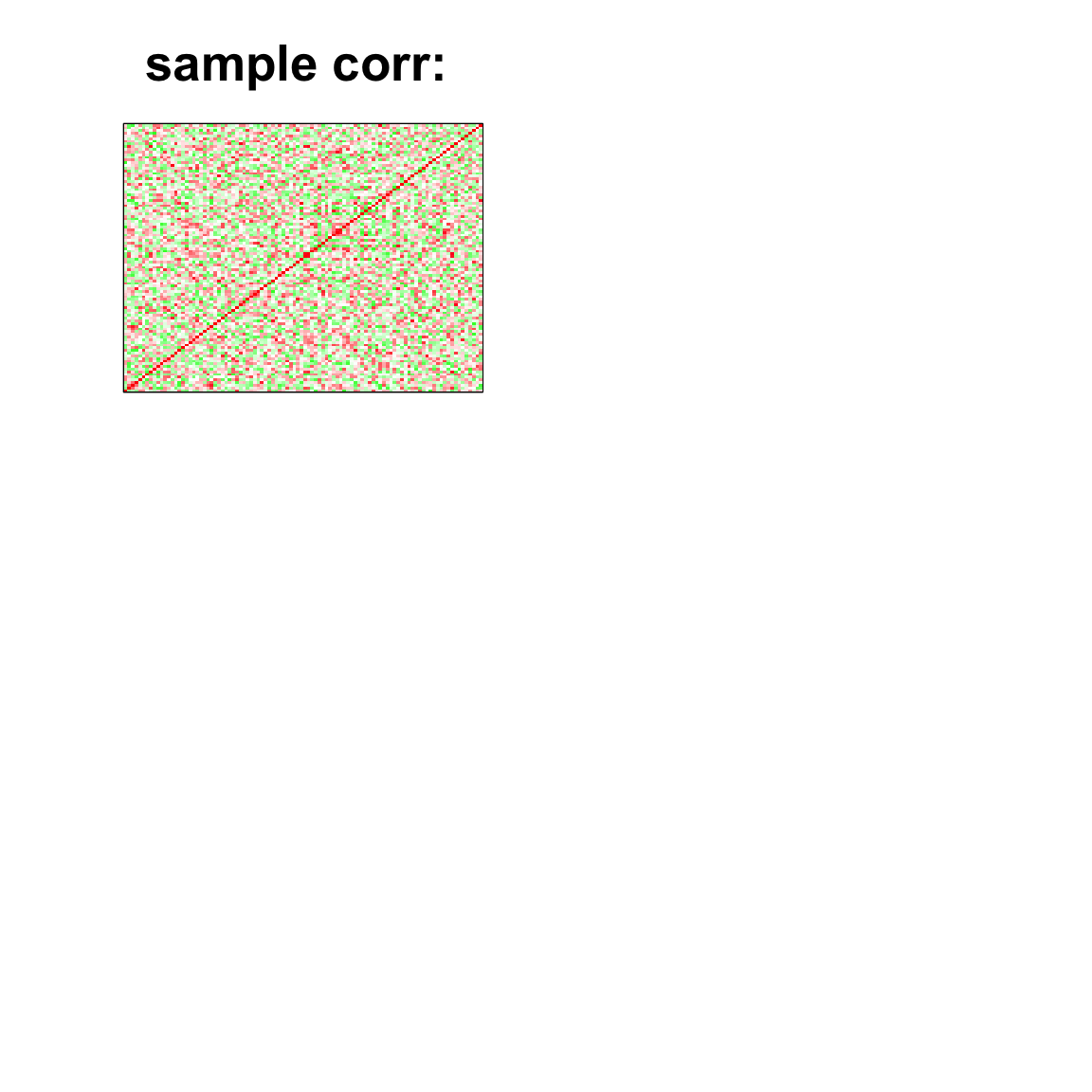
par(mfrow=c(2,2))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(cov_mat)),
col=col, main=paste0("sample corr: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
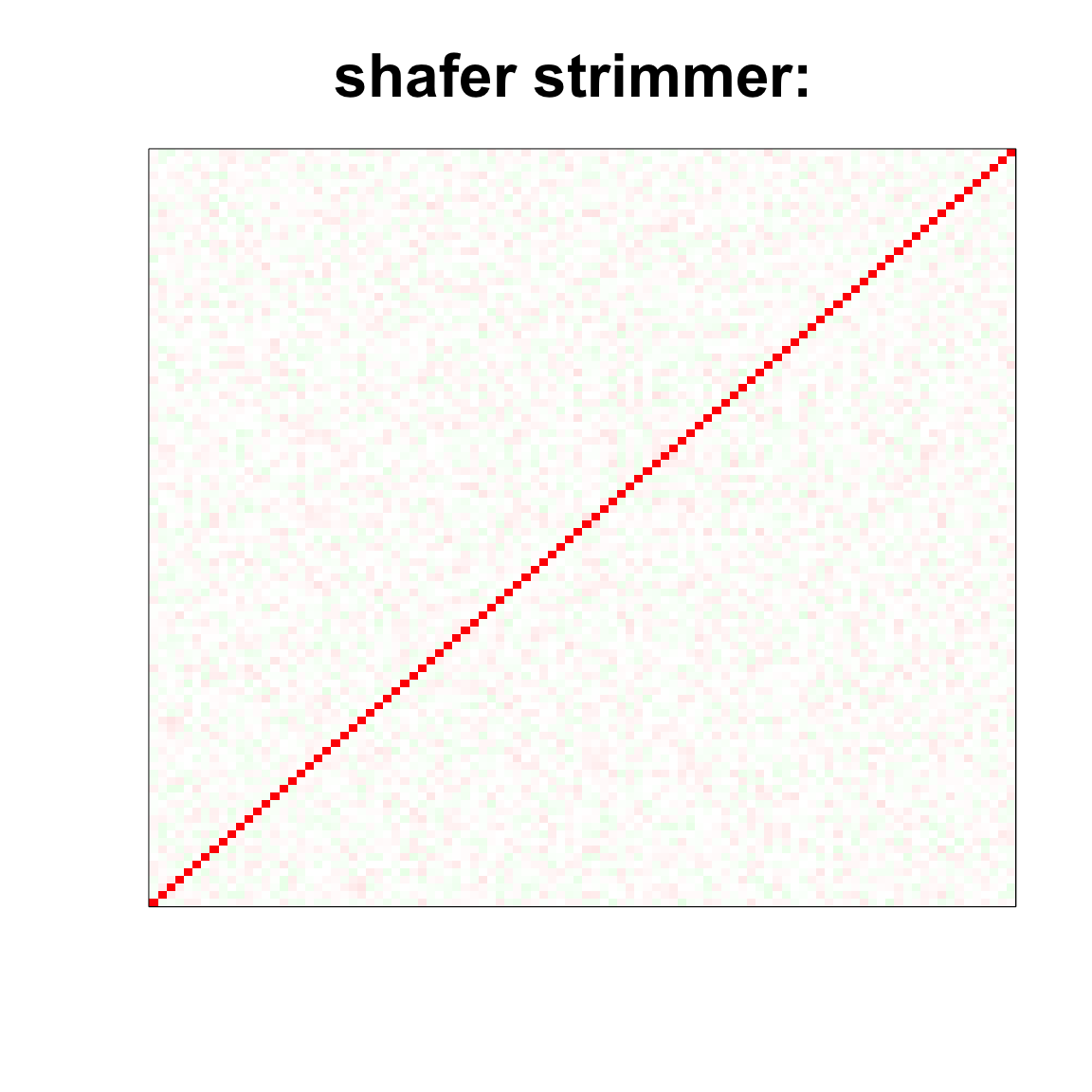
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(strimmer_sample)),
col=col, main=paste0("shafer strimmer: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
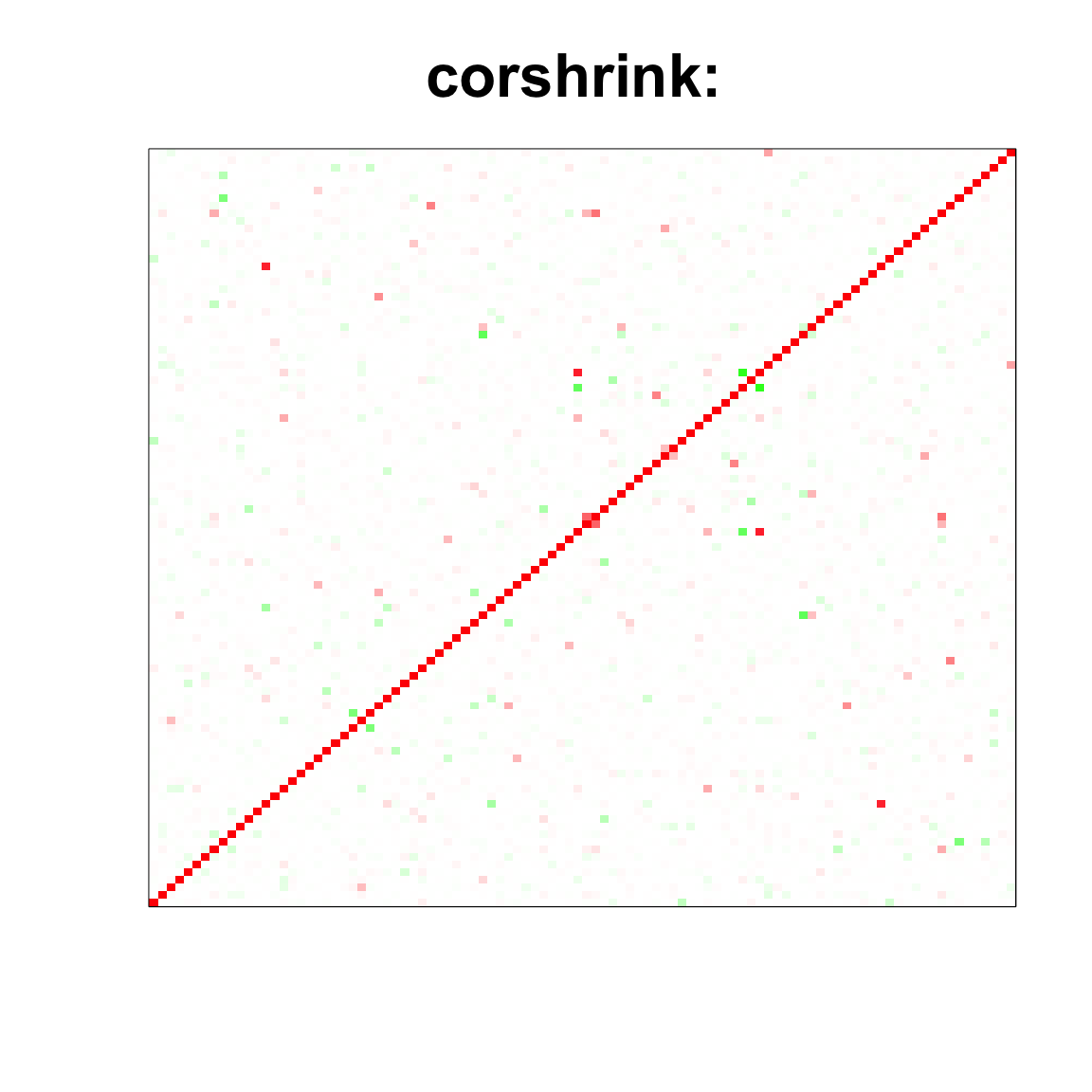
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov_sample_ML$ash_cor_only),
col=col, main=paste0("corshrink: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(glasso_sample_005$w)),
col=col, main=paste0("glasso 0.05: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(glasso_sample_05$w)),
col=col, main=paste0("glasso 0.5: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(spcor_mat),
col=col, main=paste0("spcov:", expression(lambda), "=", lam), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
n = 50, P=100
n <- 50
P <- 100
ll <- DM_graph(n,P,g_type = "cluster", para = 5)
## Generating data from the multivariate normal distribution with the cluster graph structure....
## done.
data <- rbind(ll$Xtrain, ll$Xtest)
Sigma <- ll$Sigma
corSigma <- cov2cor(Sigma)
Pat <- matrix(1, P, P)
diag(Pat) <- 0
lam <- 0.06
step.size <- 100
tol <- 1e-4
covmat <- cov(data)
mm <- spcov(Sigma=covmat+0.1*diag(1,P), S=covmat+0.1*diag(1,P), lambda=lam * Pat,step.size=step.size, n.inner.steps=200, thr.inner=0, tol.outer=tol, trace=1)
## ---
## using Nesterov, backtracking line search
## ---
## objective: 76.26152
## step size given to GGDescent/Nesterov: 100
## objective: 41.8605 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 20
## step size given to GGDescent/Nesterov: 20
## objective: 40.99096 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 4
## step size given to GGDescent/Nesterov: 4
## objective: 40.93208 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 0.8
## step size given to GGDescent/Nesterov: 0.8
## objective: 40.92464 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 0.16
## step size given to GGDescent/Nesterov: 0.16
## objective: 40.92308 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 0.032
## step size given to GGDescent/Nesterov: 0.032
## objective: 40.92265 ( 200 iterations, max step size: 0.0064 )
## step size given to GGDescent/Nesterov: 0.032
## objective: 40.92252 ( 200 iterations, max step size: 0.0064 )
## step size given to GGDescent/Nesterov: 0.032
## objective: 40.92248 ( 200 iterations, max step size: 0.0064 )
## MM converged in 8 steps!
spcor_mat <- cov2cor(mm$Sigma)
#devtools::install_github("kkdey/CorShrink")
#library(CorShrink)
#sessionInfo()
cov_mat <- cov(data);
system.time(strimmer_sample <- corpcor::cov.shrink(data))
## Estimating optimal shrinkage intensity lambda.var (variance vector): 0.9144
##
## Estimating optimal shrinkage intensity lambda (correlation matrix): 0.8058
## user system elapsed
## 0.005 0.000 0.005
system.time(glasso_sample_005 <- glasso::glasso(cov_mat, rho = 0.05))
## user system elapsed
## 0.157 0.000 0.157
system.time(glasso_sample_05 <- glasso::glasso(cov_mat, rho = 0.5))
## user system elapsed
## 0.002 0.000 0.001
system.time(glasso_sample_1 <- glasso::glasso(cov_mat, rho = 1))
## user system elapsed
## 0.001 0.000 0.002
system.time(glasso_sample_10 <- glasso::glasso(cov_mat, rho = 10))
## user system elapsed
## 0.001 0.000 0.001
system.time(cov_sample_ML <- CorShrinkMatrix(cov2cor(cov_mat), nsamp = matrix(n, P, P), ash.control = list(mixcompdist = "normal", nullweight = 10)))
## ash cor only and ash cor PD matrices are different
## user system elapsed
## 2.803 0.068 2.891
par(mfrow = c(2,2))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
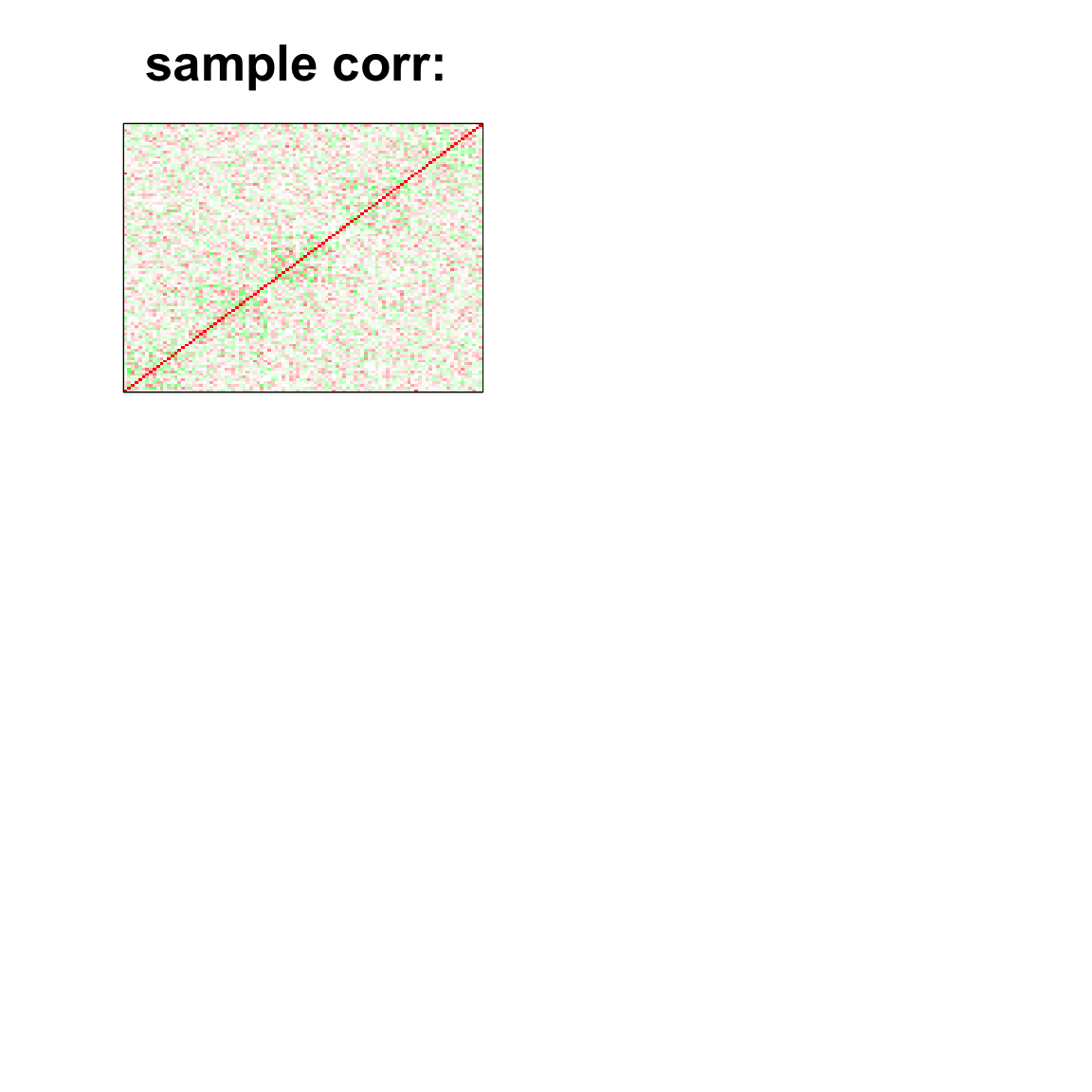
image(as.matrix(cov2cor(cov_mat)),
col=col, main=paste0("sample corr: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
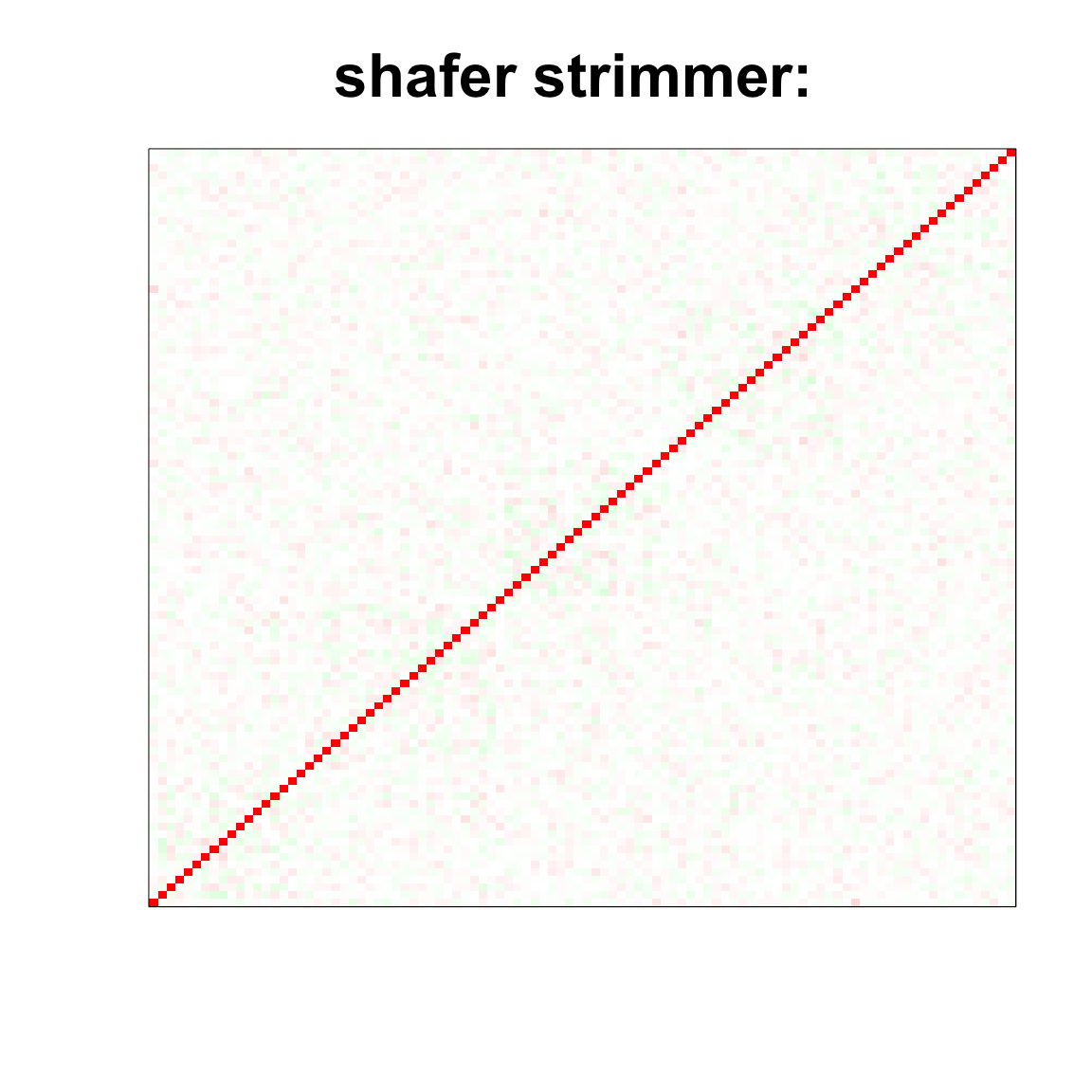
image(as.matrix(cov2cor(strimmer_sample)),
col=col, main=paste0("shafer strimmer: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
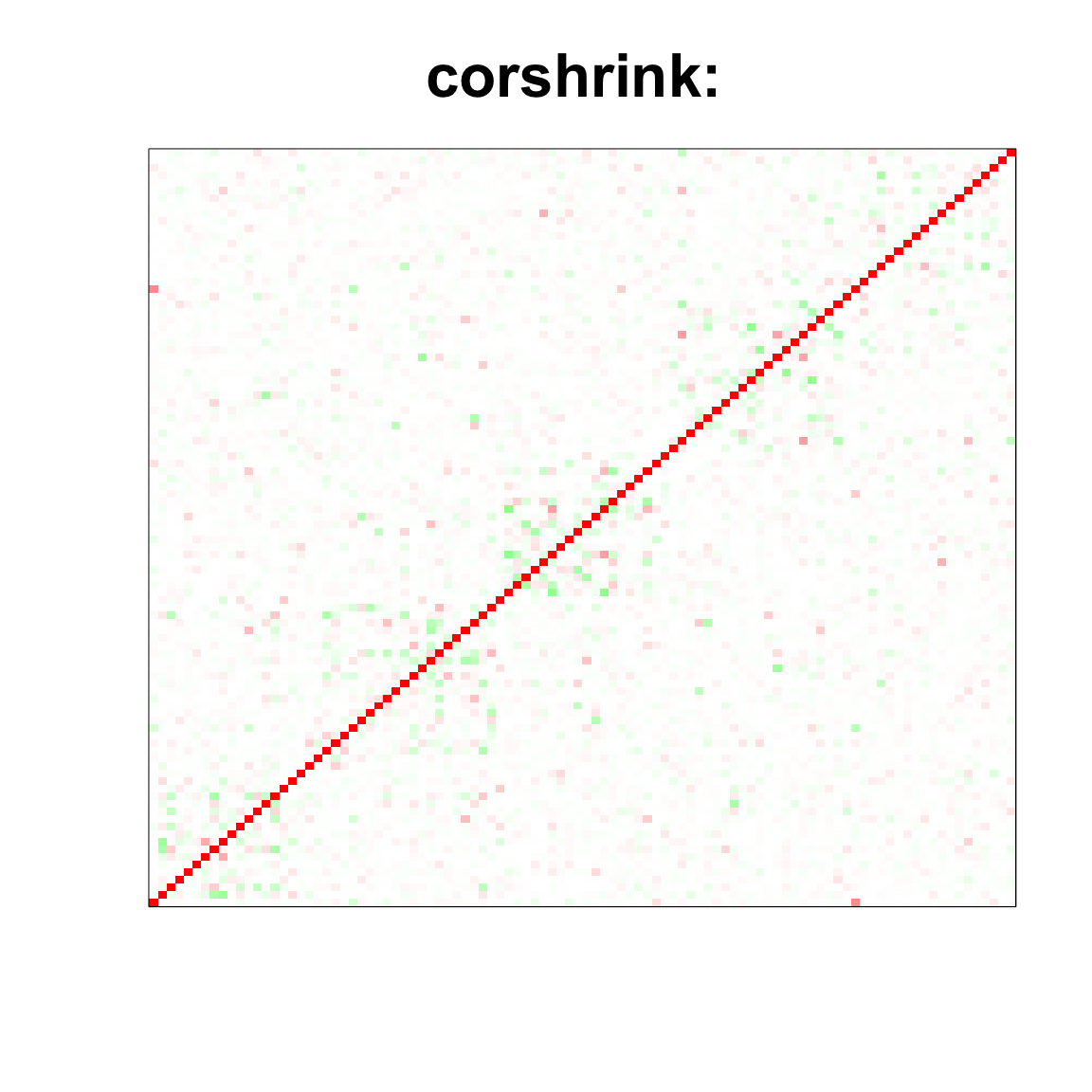
image(as.matrix(cov_sample_ML$ash_cor_only),
col=col, main=paste0("corshrink: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
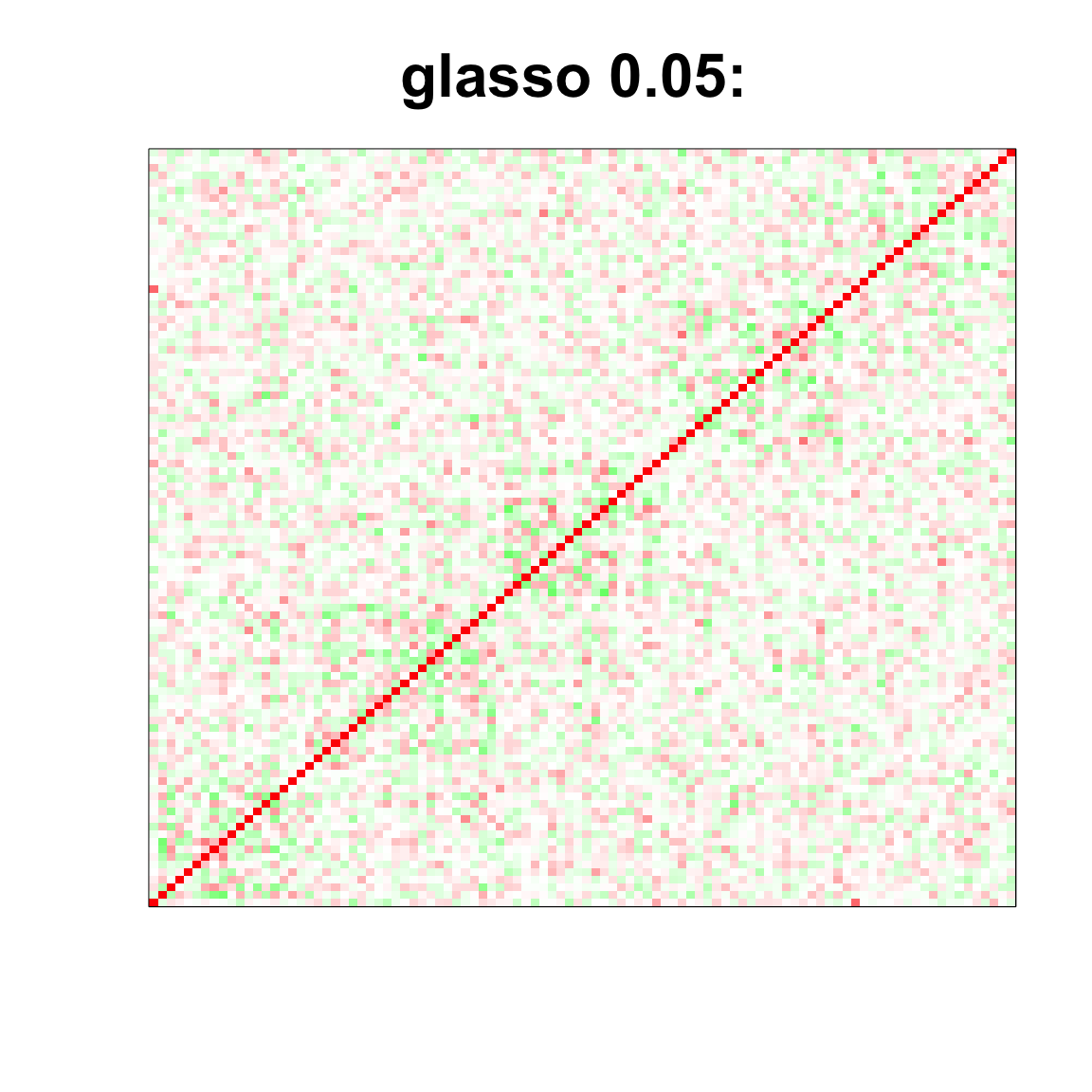
image(as.matrix(cov2cor(glasso_sample_005$w)),
col=col, main=paste0("glasso 0.05: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(glasso_sample_05$w)),
col=col, main=paste0("glasso 0.5: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
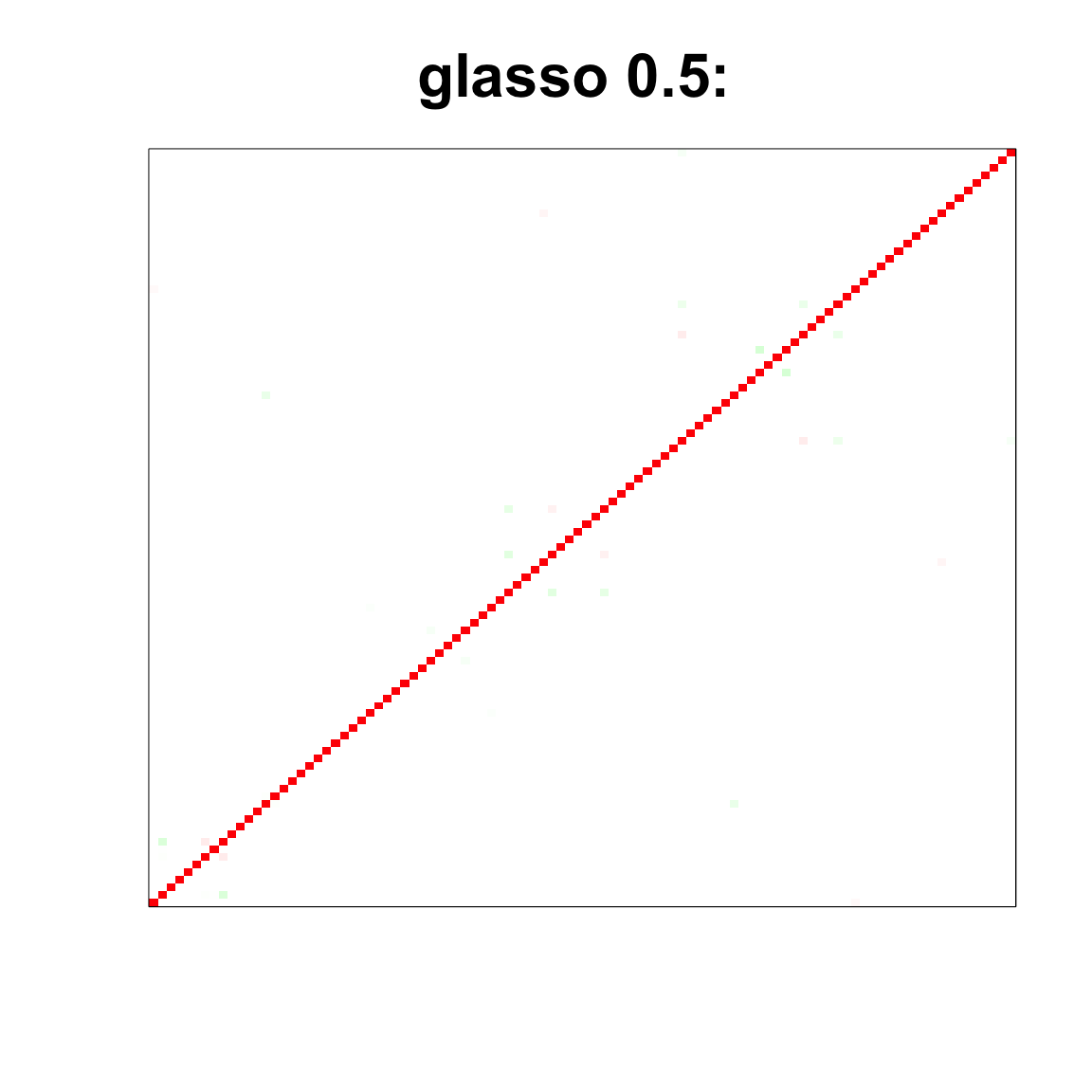
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(spcor_mat),
col=col, main=paste0("spcov:", expression(lambda), "=", lam), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
n = 5, P=100
n <- 5
P <- 100
ll <- DM_graph(n,P,g_type = "cluster", para = 5)
## Generating data from the multivariate normal distribution with the cluster graph structure....
## done.
data <- rbind(ll$Xtrain, ll$Xtest)
Sigma <- ll$Sigma
corSigma <- cov2cor(Sigma)
Pat <- matrix(1, P, P)
diag(Pat) <- 0
lam <- 0.06
step.size <- 100
tol <- 1e-4
covmat <- cov(data)
mm <- spcov(Sigma=covmat+0.1*diag(1,P), S=covmat+0.1*diag(1,P), lambda=lam * Pat,step.size=step.size, n.inner.steps=200, thr.inner=0, tol.outer=tol, trace=1)
## ---
## using Nesterov, backtracking line search
## ---
## objective: 129.7098
## step size given to GGDescent/Nesterov: 100
## objective: -59.88169 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 20
## step size given to GGDescent/Nesterov: 20
## objective: -61.31028 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 4
## step size given to GGDescent/Nesterov: 4
## objective: -61.31996 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.8
## step size given to GGDescent/Nesterov: 0.8
## objective: -61.32144 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.16
## step size given to GGDescent/Nesterov: 0.16
## objective: -61.32183 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.032
## step size given to GGDescent/Nesterov: 0.032
## objective: -61.32194 ( 200 iterations, max step size: 0.00128 )
## step size given to GGDescent/Nesterov: 0.032
## objective: -61.32197 ( 200 iterations, max step size: 0.00128 )
## MM converged in 7 steps!
spcor_mat <- cov2cor(mm$Sigma)
#devtools::install_github("kkdey/CorShrink")
#library(CorShrink)
#sessionInfo()
cov_mat <- cov(data);
system.time(strimmer_sample <- corpcor::cov.shrink(data))
## Estimating optimal shrinkage intensity lambda.var (variance vector): 0.4911
##
## Estimating optimal shrinkage intensity lambda (correlation matrix): 0.7269
## user system elapsed
## 0.002 0.000 0.002
system.time(glasso_sample_005 <- glasso::glasso(cov_mat, rho = 0.05))
## user system elapsed
## 0.615 0.002 0.620
system.time(glasso_sample_05 <- glasso::glasso(cov_mat, rho = 0.5))
## user system elapsed
## 0.061 0.000 0.062
system.time(glasso_sample_1 <- glasso::glasso(cov_mat, rho = 1))
## user system elapsed
## 0.016 0.000 0.017
system.time(glasso_sample_10 <- glasso::glasso(cov_mat, rho = 10))
## user system elapsed
## 0.001 0.000 0.002
system.time(cov_sample_ML <- CorShrinkMatrix(cov2cor(cov_mat), nsamp = matrix(n, P, P), ash.control = list(mixcompdist = "normal", nullweight = 10)))
## ash cor only and ash cor PD matrices are different
## user system elapsed
## 2.043 0.048 2.100
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(cov_mat)),
col=col, main=paste0("sample corr: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
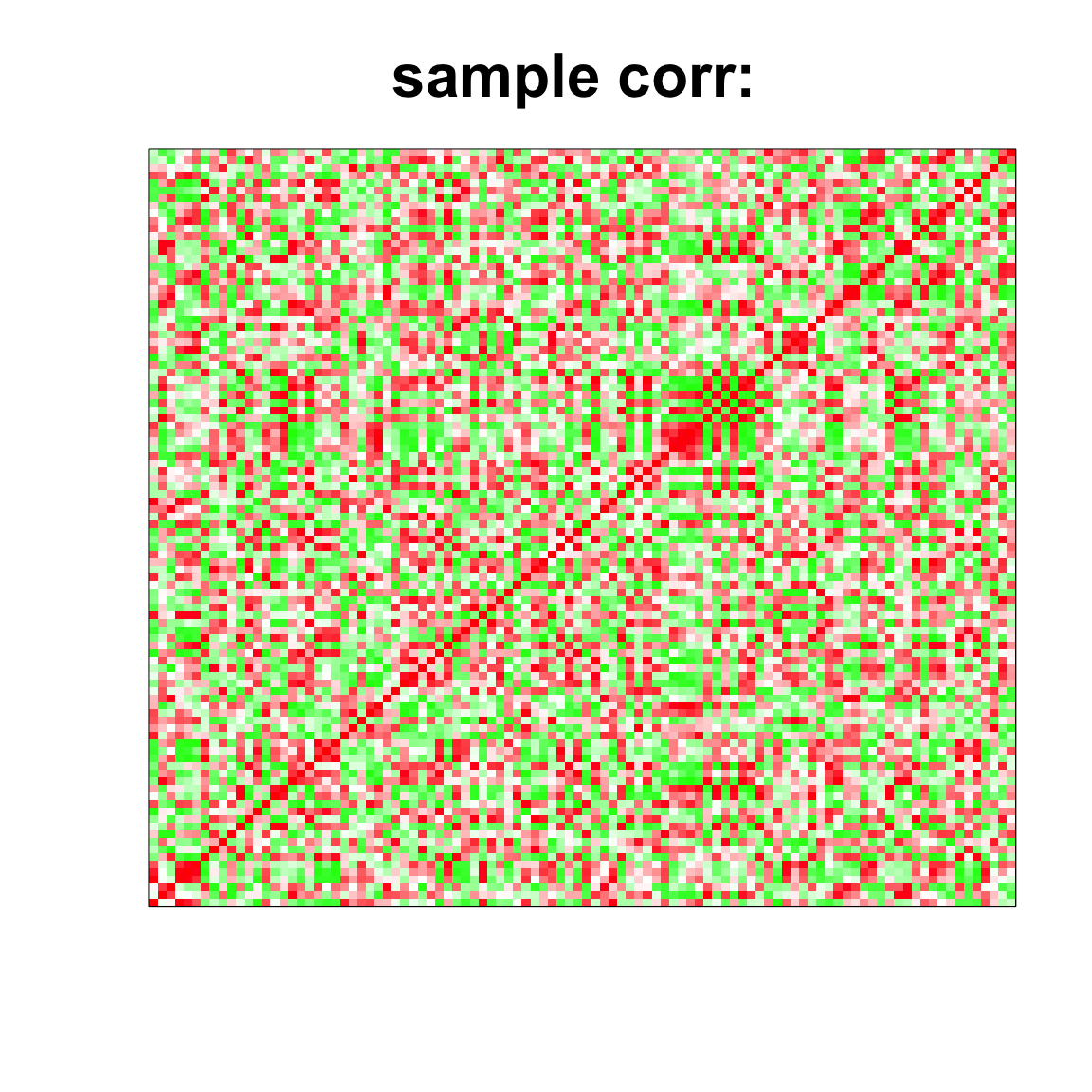
par(mfrow=c(2,2))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(strimmer_sample)),
col=col, main=paste0("shafer strimmer: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov_sample_ML$ash_cor_only),
col=col, main=paste0("corshrink: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(glasso_sample_005$w)),
col=col, main=paste0("glasso 0.05: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(glasso_sample_05$w)),
col=col, main=paste0("glasso 0.5: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
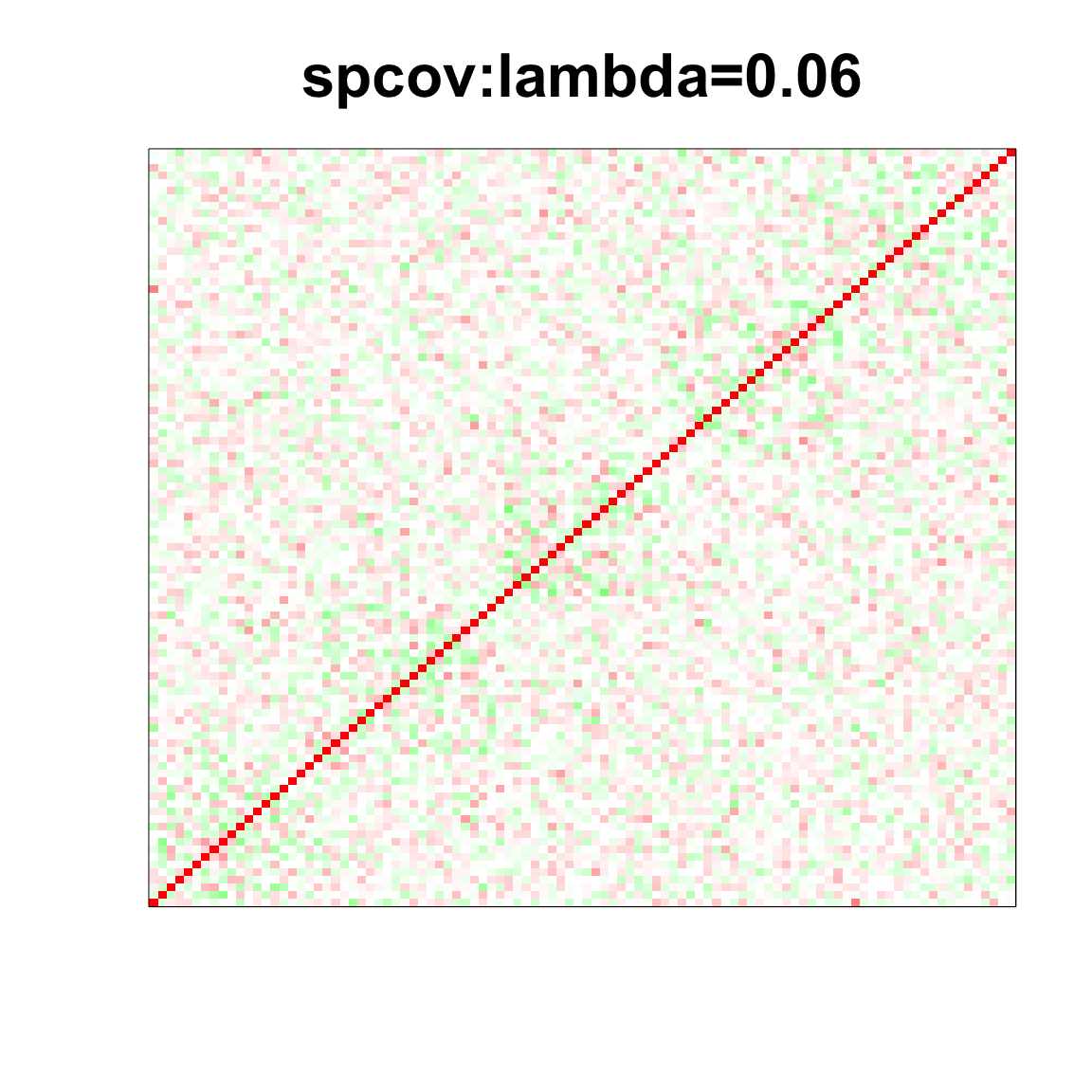
image(as.matrix(spcor_mat),
col=col, main=paste0("spcov:", expression(lambda), "=", lam), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
DM graph (dense Edors Renyi graph)
n = 10, P=100
n <- 10
P <- 100
ll <- DM_graph(n,P, para = 0.5)
## Generating data from the multivariate normal distribution with the random graph structure....
## done.
data <- rbind(ll$Xtrain, ll$Xtest)
Sigma <- ll$Sigma
corSigma <- cov2cor(Sigma)
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(corSigma),
col=col, main=paste0("pop corr: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
Pat <- matrix(1, P, P)
diag(Pat) <- 0
lam <- 0.06
step.size <- 100
tol <- 1e-4
covmat <- cov(data)
mm <- spcov(Sigma=covmat+0.1*diag(1,P), S=covmat+0.1*diag(1,P), lambda=lam * Pat,step.size=step.size, n.inner.steps=200, thr.inner=0, tol.outer=tol, trace=1)
## ---
## using Nesterov, backtracking line search
## ---
## objective: 104.3729
## step size given to GGDescent/Nesterov: 100
## objective: -31.73461 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 20
## step size given to GGDescent/Nesterov: 20
## objective: -33.7312 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 4
## step size given to GGDescent/Nesterov: 4
## objective: -33.75088 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.8
## step size given to GGDescent/Nesterov: 0.8
## objective: -33.75285 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.16
## step size given to GGDescent/Nesterov: 0.16
## objective: -33.75331 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.032
## step size given to GGDescent/Nesterov: 0.032
## objective: -33.75343 ( 200 iterations, max step size: 0.00128 )
## step size given to GGDescent/Nesterov: 0.032
## objective: -33.75347 ( 200 iterations, max step size: 0.00128 )
## MM converged in 7 steps!
spcor_mat <- cov2cor(mm$Sigma)
#devtools::install_github("kkdey/CorShrink")
#library(CorShrink)
#sessionInfo()
cov_mat <- cov(data);
system.time(strimmer_sample <- corpcor::cov.shrink(data))
## Estimating optimal shrinkage intensity lambda.var (variance vector): 1
##
## Estimating optimal shrinkage intensity lambda (correlation matrix): 0.8107
## user system elapsed
## 0.002 0.000 0.002
system.time(glasso_sample_005 <- glasso::glasso(cov_mat, rho = 0.05))
## user system elapsed
## 0.434 0.001 0.436
system.time(glasso_sample_05 <- glasso::glasso(cov_mat, rho = 0.5))
## user system elapsed
## 0.044 0.000 0.044
system.time(glasso_sample_1 <- glasso::glasso(cov_mat, rho = 1))
## user system elapsed
## 0.004 0.000 0.004
system.time(glasso_sample_10 <- glasso::glasso(cov_mat, rho = 10))
## user system elapsed
## 0.001 0.000 0.001
system.time(cov_sample_ML <- CorShrinkMatrix(cov2cor(cov_mat), nsamp = matrix(n, P, P), ash.control = list(mixcompdist = "normal", nullweight = 10)))
## ash cor only and ash cor PD matrices are different
## user system elapsed
## 4.921 0.087 5.031
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
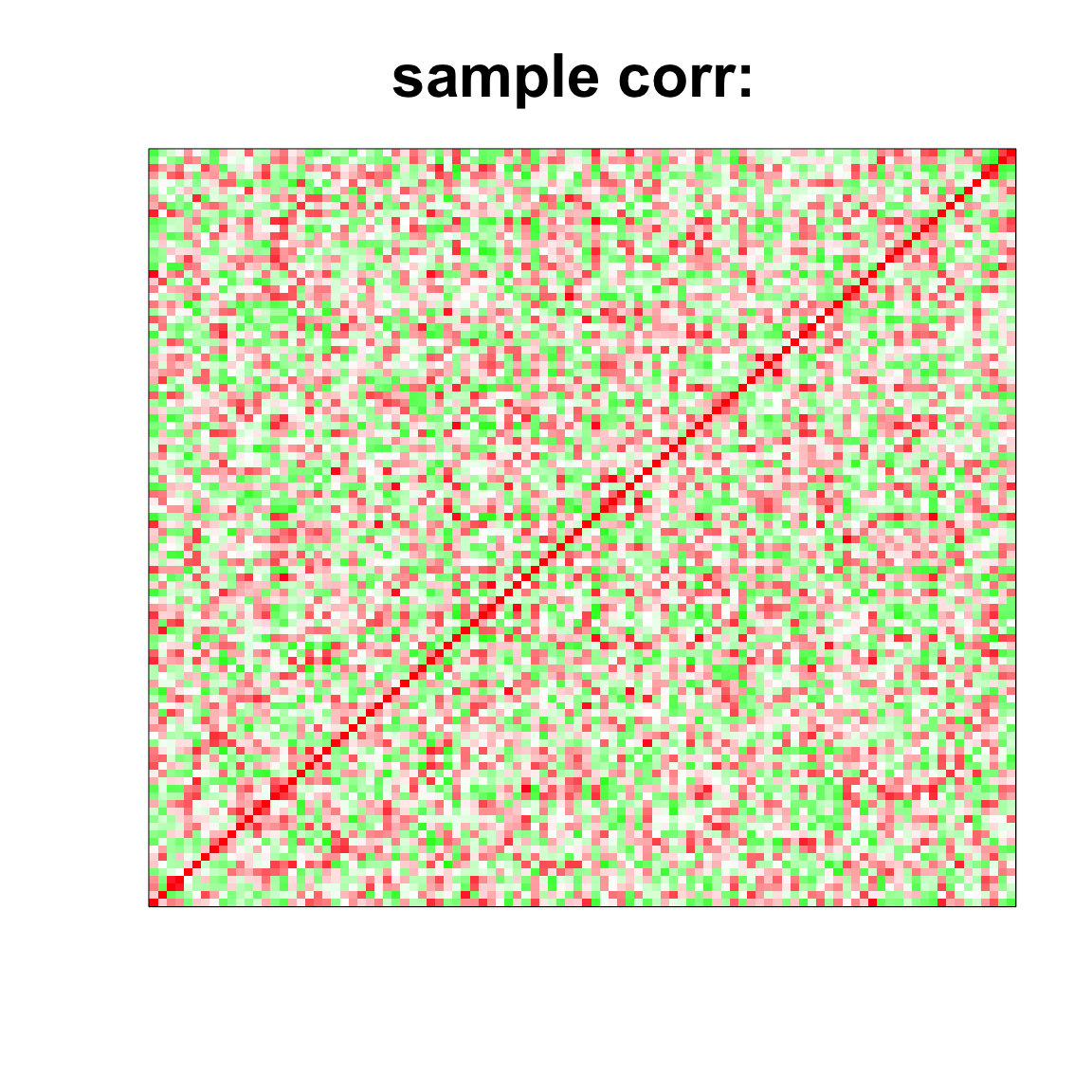
image(as.matrix(cov2cor(cov_mat)),
col=col, main=paste0("sample corr: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
par(mfrow = c(2,2))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
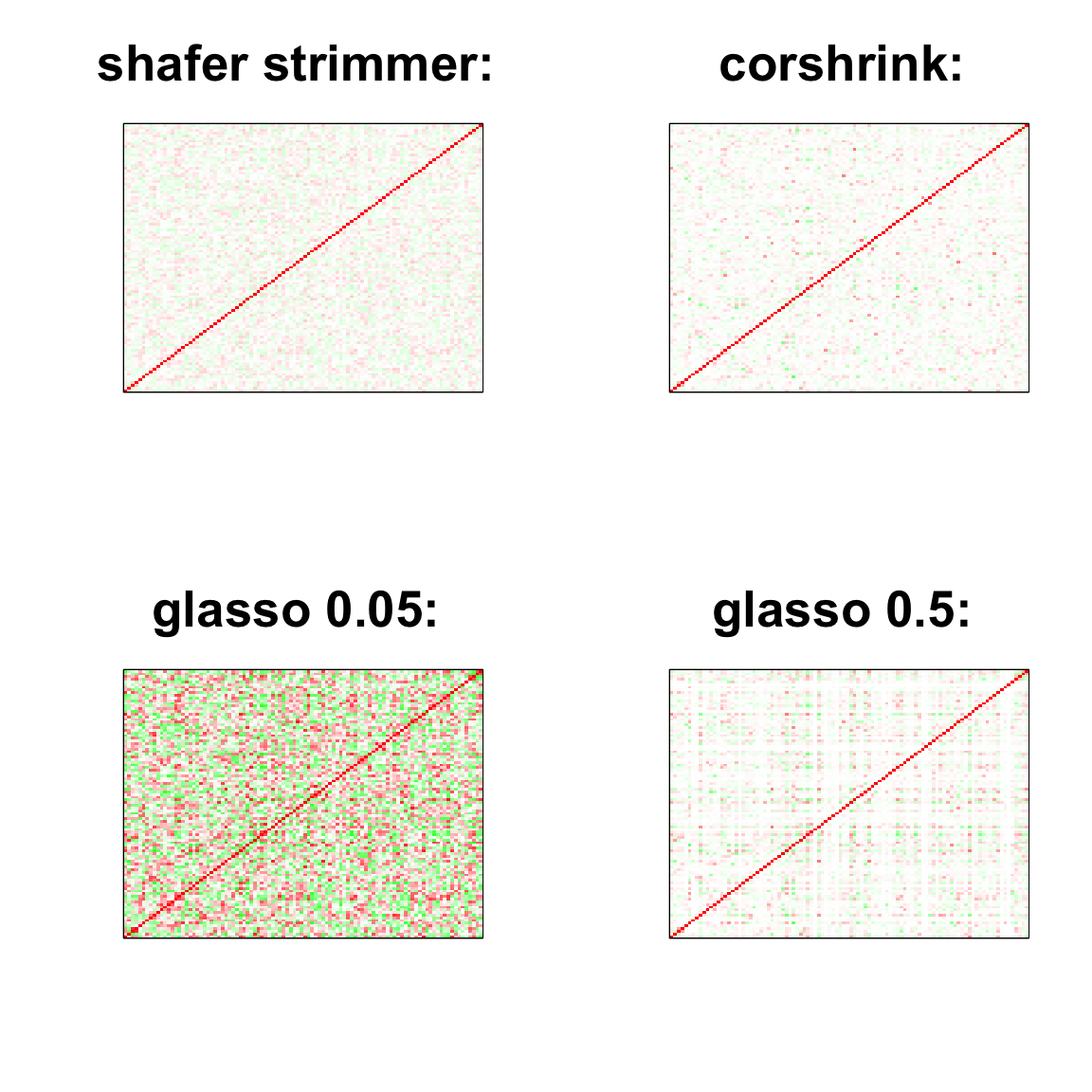
image(as.matrix(cov2cor(strimmer_sample)),
col=col, main=paste0("shafer strimmer: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov_sample_ML$ash_cor_only),
col=col, main=paste0("corshrink: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(glasso_sample_005$w)),
col=col, main=paste0("glasso 0.05: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(glasso_sample_05$w)),
col=col, main=paste0("glasso 0.5: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
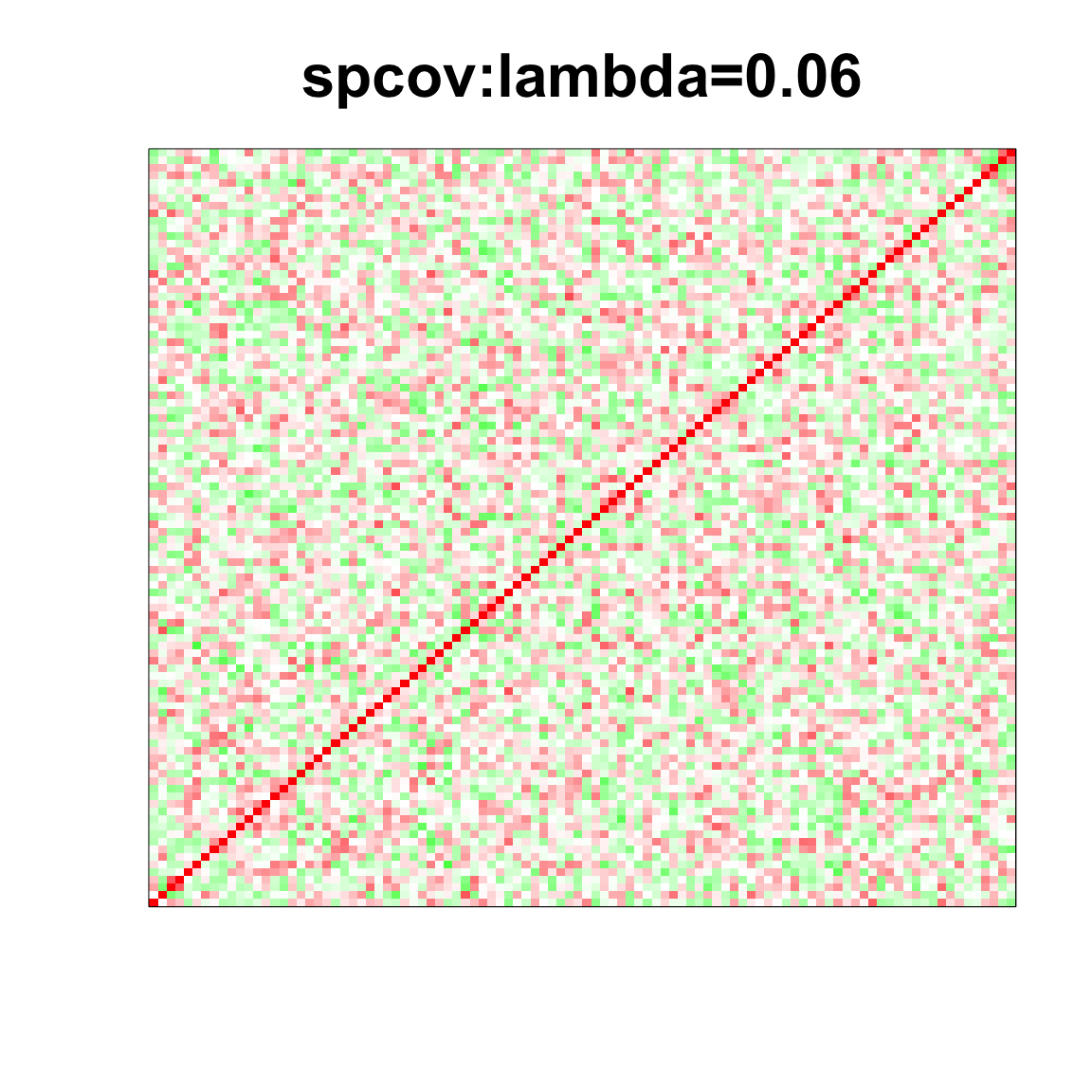
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(spcor_mat),
col=col, main=paste0("spcov:", expression(lambda), "=", lam), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
n = 50, P=100
n <- 50
P <- 100
ll <- DM_graph(n,P, para = 0.5)
## Generating data from the multivariate normal distribution with the random graph structure....
## done.
data <- rbind(ll$Xtrain, ll$Xtest)
Sigma <- ll$Sigma
corSigma <- cov2cor(Sigma)
Pat <- matrix(1, P, P)
diag(Pat) <- 0
lam <- 0.06
step.size <- 100
tol <- 1e-4
covmat <- cov(data)
mm <- spcov(Sigma=covmat+0.1*diag(1,P), S=covmat+0.1*diag(1,P), lambda=lam * Pat,step.size=step.size, n.inner.steps=200, thr.inner=0, tol.outer=tol, trace=1)
## ---
## using Nesterov, backtracking line search
## ---
## objective: 83.33297
## step size given to GGDescent/Nesterov: 100
## objective: 38.95256 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 20
## step size given to GGDescent/Nesterov: 20
## objective: 38.11965 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 4
## step size given to GGDescent/Nesterov: 4
## objective: 38.06355 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 0.8
## step size given to GGDescent/Nesterov: 0.8
## objective: 38.05627 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 0.16
## step size given to GGDescent/Nesterov: 0.16
## objective: 38.05473 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 0.032
## step size given to GGDescent/Nesterov: 0.032
## objective: 38.05431 ( 200 iterations, max step size: 0.0064 )
## step size given to GGDescent/Nesterov: 0.032
## objective: 38.05418 ( 200 iterations, max step size: 0.0064 )
## step size given to GGDescent/Nesterov: 0.032
## objective: 38.05414 ( 200 iterations, max step size: 0.0064 )
## MM converged in 8 steps!
spcor_mat <- cov2cor(mm$Sigma)
#devtools::install_github("kkdey/CorShrink")
#library(CorShrink)
#sessionInfo()
cov_mat <- cov(data);
system.time(strimmer_sample <- corpcor::cov.shrink(data))
## Estimating optimal shrinkage intensity lambda.var (variance vector): 1
##
## Estimating optimal shrinkage intensity lambda (correlation matrix): 0.6314
## user system elapsed
## 0.004 0.001 0.005
system.time(glasso_sample_005 <- glasso::glasso(cov_mat, rho = 0.05))
## user system elapsed
## 0.166 0.000 0.167
system.time(glasso_sample_05 <- glasso::glasso(cov_mat, rho = 0.5))
## user system elapsed
## 0.002 0.000 0.002
system.time(glasso_sample_1 <- glasso::glasso(cov_mat, rho = 1))
## user system elapsed
## 0.001 0.000 0.001
system.time(glasso_sample_10 <- glasso::glasso(cov_mat, rho = 10))
## user system elapsed
## 0.001 0.000 0.002
system.time(cov_sample_ML <- CorShrinkMatrix(cov2cor(cov_mat), nsamp = matrix(n, P, P), ash.control = list(mixcompdist = "normal", nullweight = 10)))
## ash cor only and ash cor PD matrices are different
## user system elapsed
## 6.790 0.159 6.976
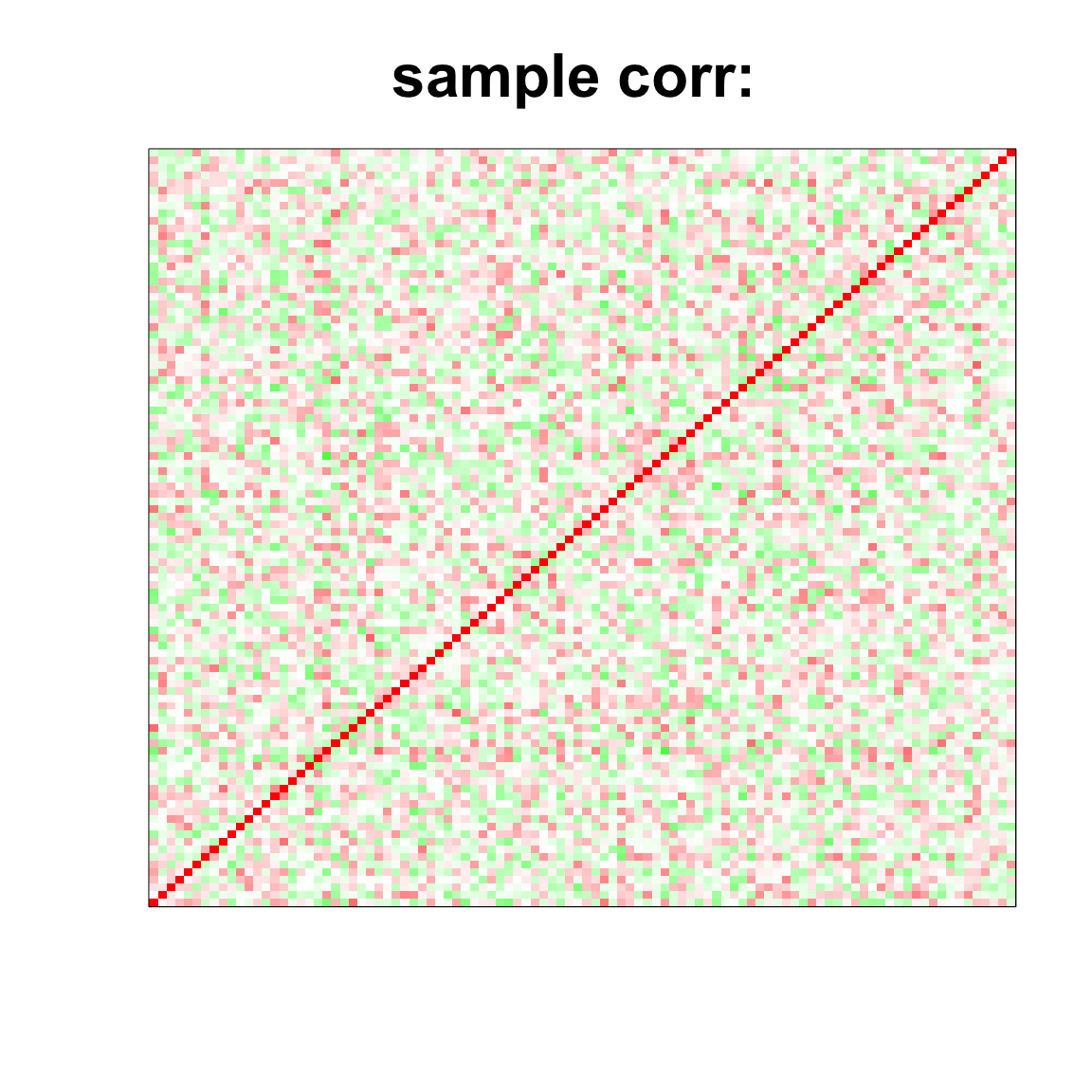
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(cov_mat)),
col=col, main=paste0("sample corr: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
par(mfrow = c(2,2))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(strimmer_sample)),
col=col, main=paste0("shafer strimmer: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov_sample_ML$ash_cor_only),
col=col, main=paste0("corshrink: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(glasso_sample_005$w)),
col=col, main=paste0("glasso 0.05: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(glasso_sample_05$w)),
col=col, main=paste0("glasso 0.5: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
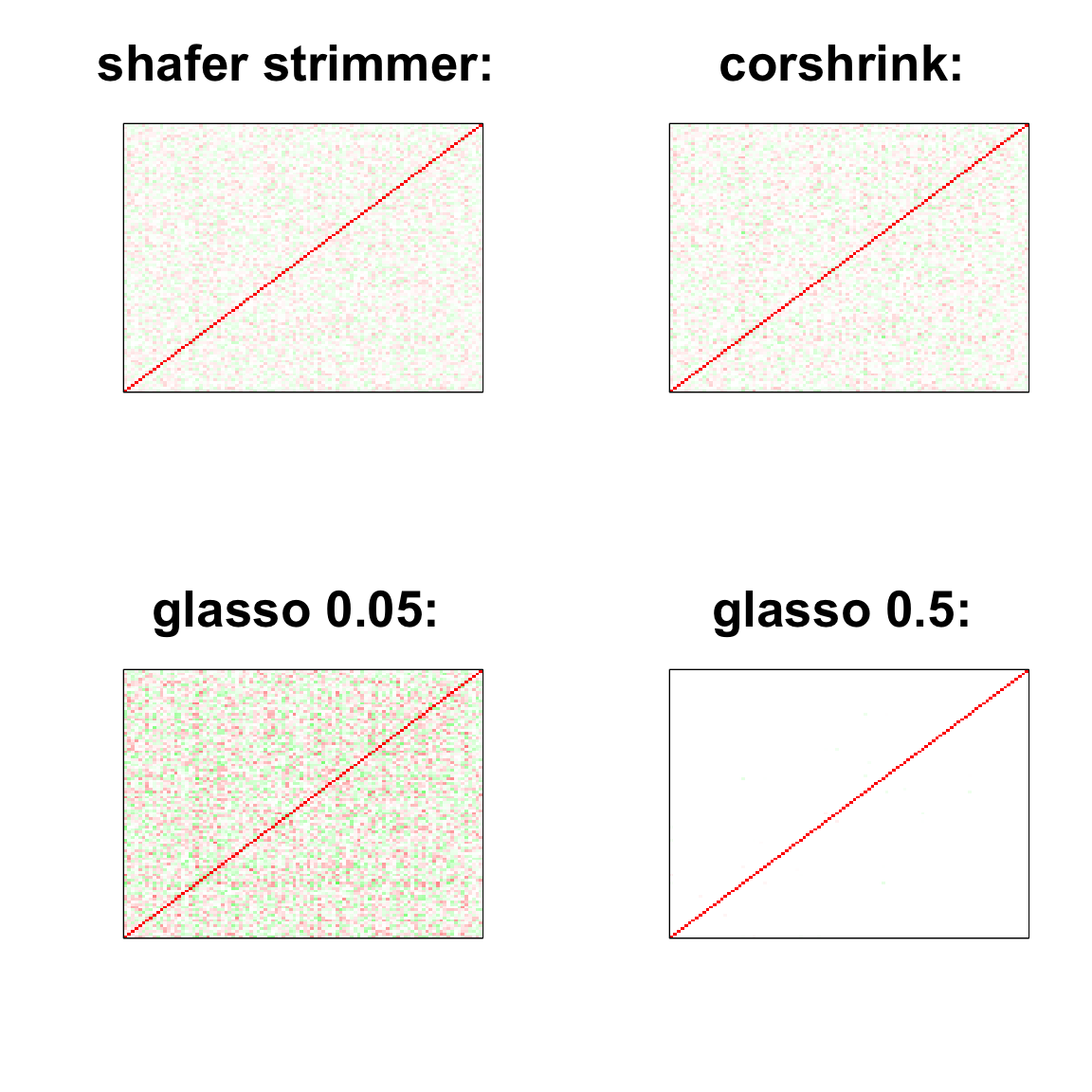
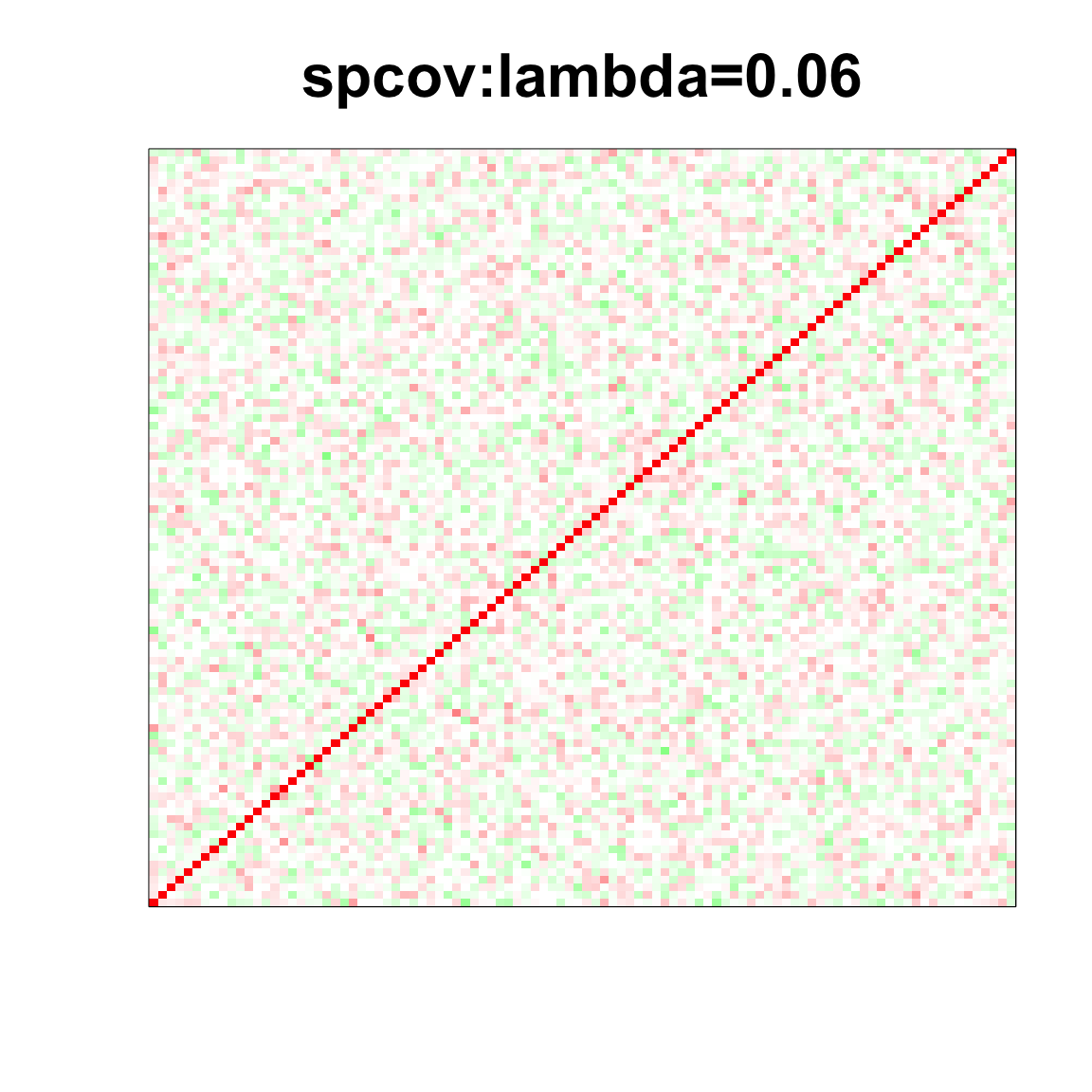
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(spcor_mat),
col=col, main=paste0("spcov:", expression(lambda), "=", lam), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
n = 5, P=100
n <- 5
P <- 100
ll <- DM_graph(n,P, para = 0.5)
## Generating data from the multivariate normal distribution with the random graph structure....
## done.
data <- rbind(ll$Xtrain, ll$Xtest)
Sigma <- ll$Sigma
corSigma <- cov2cor(Sigma)
Pat <- matrix(1, P, P)
diag(Pat) <- 0
lam <- 0.06
step.size <- 100
tol <- 1e-4
covmat <- cov(data)
mm <- spcov(Sigma=covmat+0.1*diag(1,P), S=covmat+0.1*diag(1,P), lambda=lam * Pat,step.size=step.size, n.inner.steps=200, thr.inner=0, tol.outer=tol, trace=1)
## ---
## using Nesterov, backtracking line search
## ---
## objective: 139.4486
## step size given to GGDescent/Nesterov: 100
## objective: -59.01845 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 20
## step size given to GGDescent/Nesterov: 20
## objective: -60.15098 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 4
## step size given to GGDescent/Nesterov: 4
## objective: -60.15814 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.8
## step size given to GGDescent/Nesterov: 0.8
## objective: -60.15961 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.16
## step size given to GGDescent/Nesterov: 0.16
## objective: -60.16001 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.032
## step size given to GGDescent/Nesterov: 0.032
## objective: -60.16012 ( 200 iterations, max step size: 0.00128 )
## step size given to GGDescent/Nesterov: 0.032
## objective: -60.16015 ( 200 iterations, max step size: 0.00128 )
## MM converged in 7 steps!
spcor_mat <- cov2cor(mm$Sigma)
#devtools::install_github("kkdey/CorShrink")
#library(CorShrink)
#sessionInfo()
cov_mat <- cov(data);
system.time(strimmer_sample <- corpcor::cov.shrink(data))
## Estimating optimal shrinkage intensity lambda.var (variance vector): 0.8093
##
## Estimating optimal shrinkage intensity lambda (correlation matrix): 0.7321
## user system elapsed
## 0.002 0.000 0.002
system.time(glasso_sample_005 <- glasso::glasso(cov_mat, rho = 0.05))
## user system elapsed
## 0.628 0.001 0.631
system.time(glasso_sample_05 <- glasso::glasso(cov_mat, rho = 0.5))
## user system elapsed
## 0.061 0.000 0.062
system.time(glasso_sample_1 <- glasso::glasso(cov_mat, rho = 1))
## user system elapsed
## 0.014 0.000 0.015
system.time(glasso_sample_10 <- glasso::glasso(cov_mat, rho = 10))
## user system elapsed
## 0.002 0.000 0.001
system.time(cov_sample_ML <- CorShrinkMatrix(cov2cor(cov_mat), nsamp = matrix(n, P, P), ash.control = list(mixcompdist = "normal", nullweight = 10)))
## ash cor only and ash cor PD matrices are different
## user system elapsed
## 2.633 0.059 2.718
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(cov_mat)),
col=col, main=paste0("sample corr: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
par(mfrow = c(2,2))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(strimmer_sample)),
col=col, main=paste0("shafer strimmer: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov_sample_ML$ash_cor_only),
col=col, main=paste0("corshrink: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(glasso_sample_005$w)),
col=col, main=paste0("glasso 0.05: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(glasso_sample_05$w)),
col=col, main=paste0("glasso 0.5: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))

col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(spcor_mat),
col=col, main=paste0("spcov:", expression(lambda), "=", lam), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
DM graph (sparse Edors Renyi graph)
n = 10, P=100
n <- 10
P <- 100
ll <- DM_graph(n,P)
## Generating data from the multivariate normal distribution with the random graph structure....
## done.
data <- rbind(ll$Xtrain, ll$Xtest)
Sigma <- ll$Sigma
corSigma <- cov2cor(Sigma)
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(corSigma),
col=col, main=paste0("pop corr: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
Pat <- matrix(1, P, P)
diag(Pat) <- 0
lam <- 0.06
step.size <- 100
tol <- 1e-4
covmat <- cov(data)
mm <- spcov(Sigma=covmat+0.1*diag(1,P), S=covmat+0.1*diag(1,P), lambda=lam * Pat,step.size=step.size, n.inner.steps=200, thr.inner=0, tol.outer=tol, trace=1)
## ---
## using Nesterov, backtracking line search
## ---
## objective: 58.33501
## step size given to GGDescent/Nesterov: 100
## objective: -40.33554 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 20
## step size given to GGDescent/Nesterov: 20
## objective: -40.93009 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 4
## step size given to GGDescent/Nesterov: 4
## objective: -40.93965 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.8
## step size given to GGDescent/Nesterov: 0.8
## objective: -40.94105 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.16
## step size given to GGDescent/Nesterov: 0.16
## objective: -40.94141 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.032
## step size given to GGDescent/Nesterov: 0.032
## objective: -40.94151 ( 200 iterations, max step size: 0.00128 )
## MM converged in 6 steps!
spcor_mat <- cov2cor(mm$Sigma)
#devtools::install_github("kkdey/CorShrink")
#library(CorShrink)
#sessionInfo()
cov_mat <- cov(data);
system.time(strimmer_sample <- corpcor::cov.shrink(data))
## Estimating optimal shrinkage intensity lambda.var (variance vector): 0.78
##
## Estimating optimal shrinkage intensity lambda (correlation matrix): 0.9079
## user system elapsed
## 0.002 0.000 0.002
system.time(glasso_sample_005 <- glasso::glasso(cov_mat, rho = 0.05))
## user system elapsed
## 0.359 0.001 0.362
system.time(glasso_sample_05 <- glasso::glasso(cov_mat, rho = 0.5))
## user system elapsed
## 0.031 0.000 0.030
system.time(glasso_sample_1 <- glasso::glasso(cov_mat, rho = 1))
## user system elapsed
## 0.002 0.000 0.002
system.time(glasso_sample_10 <- glasso::glasso(cov_mat, rho = 10))
## user system elapsed
## 0.002 0.000 0.001
system.time(cov_sample_ML <- CorShrinkMatrix(cov2cor(cov_mat), nsamp = matrix(n, P, P), ash.control = list(mixcompdist = "normal", nullweight = 10)))
## ash cor only and ash cor PD matrices are different
## user system elapsed
## 2.292 0.052 2.351
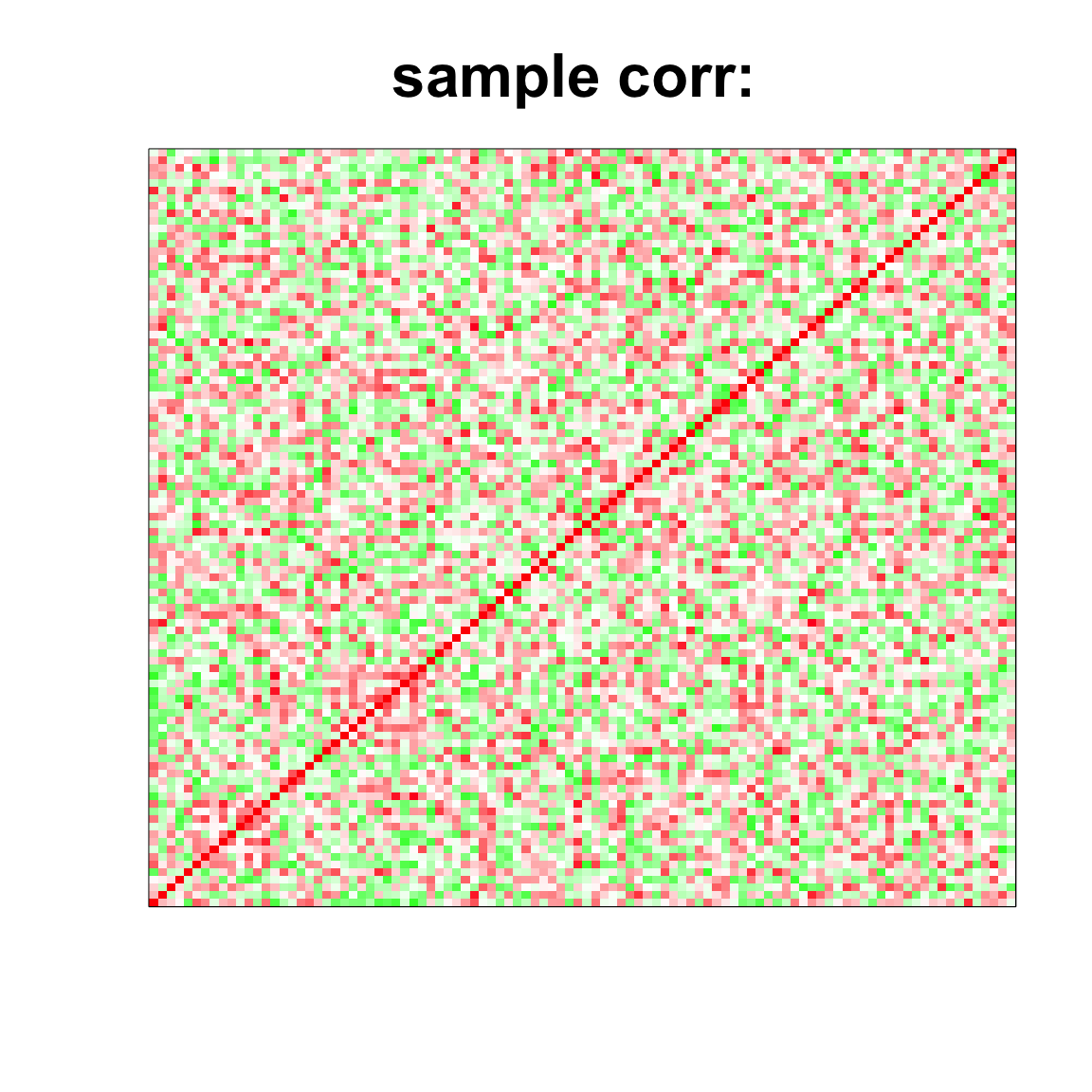
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(cov_mat)),
col=col, main=paste0("sample corr: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
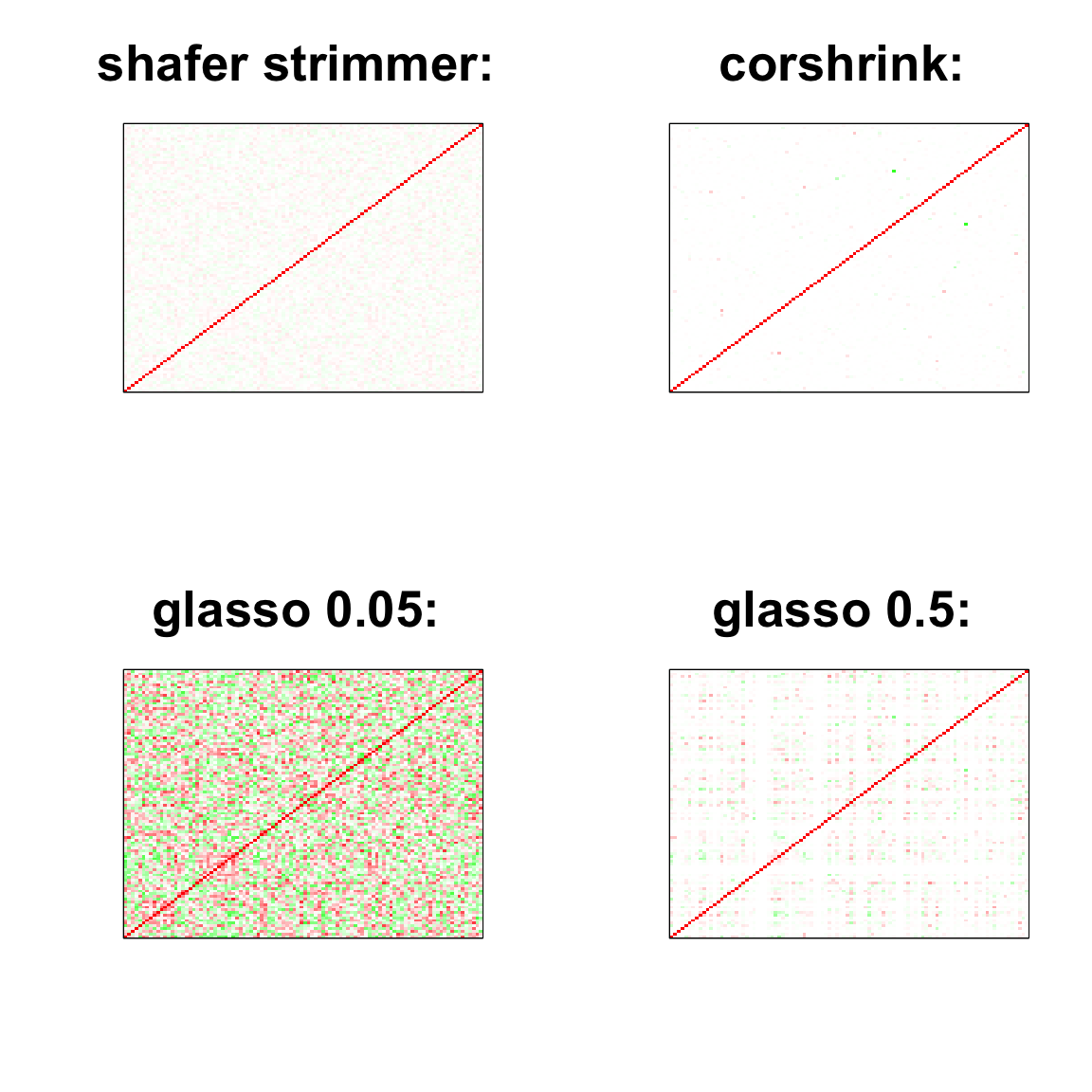
par(mfrow = c(2,2))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(strimmer_sample)),
col=col, main=paste0("shafer strimmer: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov_sample_ML$ash_cor_only),
col=col, main=paste0("corshrink: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(glasso_sample_005$w)),
col=col, main=paste0("glasso 0.05: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(glasso_sample_05$w)),
col=col, main=paste0("glasso 0.5: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
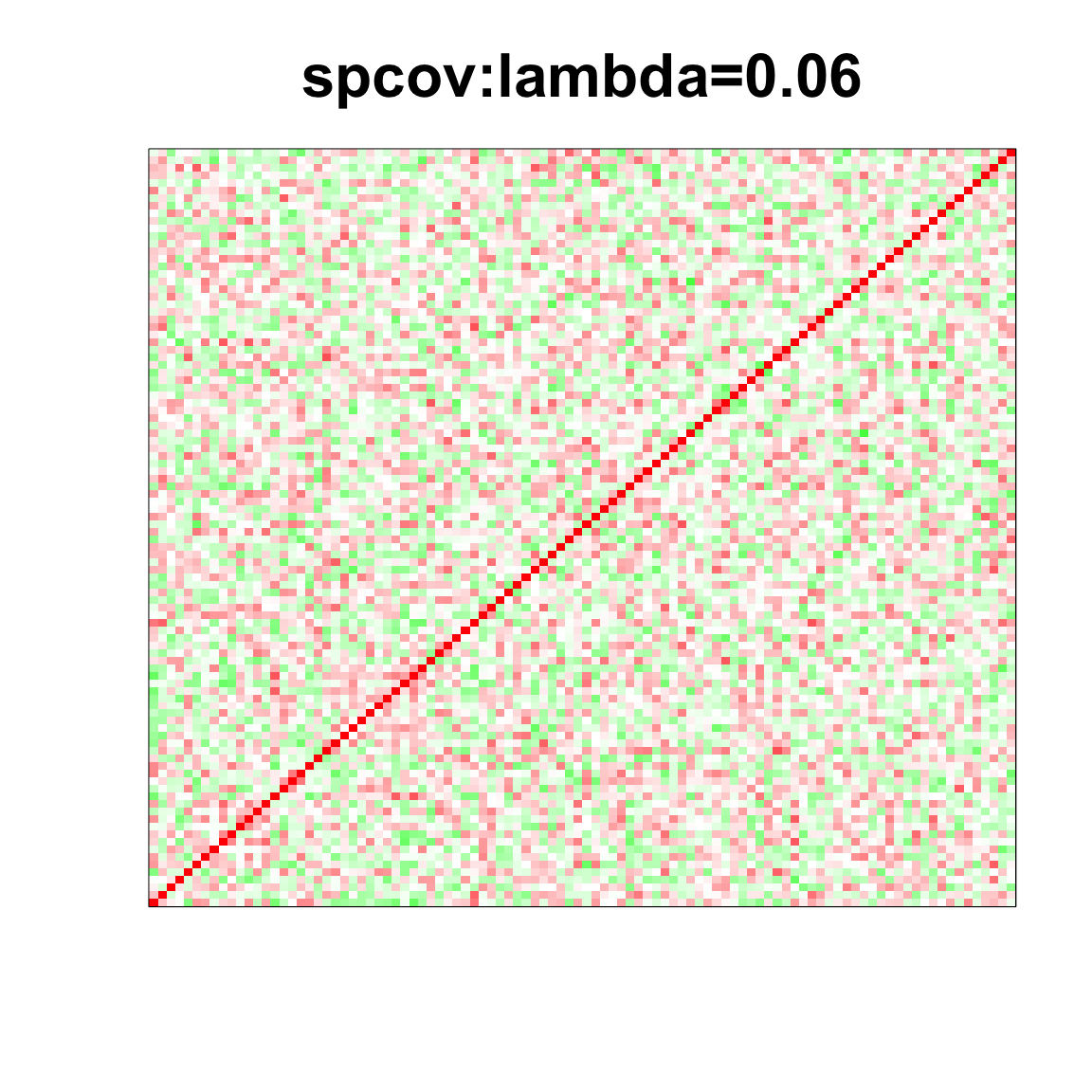
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(spcor_mat),
col=col, main=paste0("spcov:", expression(lambda), "=", lam), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
n = 50, P=100
n <- 50
P <- 100
ll <- DM_graph(n,P)
## Generating data from the multivariate normal distribution with the random graph structure....
## done.
data <- rbind(ll$Xtrain, ll$Xtest)
Sigma <- ll$Sigma
corSigma <- cov2cor(Sigma)
Pat <- matrix(1, P, P)
diag(Pat) <- 0
lam <- 0.06
step.size <- 100
tol <- 1e-4
covmat <- cov(data)
mm <- spcov(Sigma=covmat+0.1*diag(1,P), S=covmat+0.1*diag(1,P), lambda=lam * Pat,step.size=step.size, n.inner.steps=200, thr.inner=0, tol.outer=tol, trace=1)
## ---
## using Nesterov, backtracking line search
## ---
## objective: 81.1509
## step size given to GGDescent/Nesterov: 100
## objective: 45.18801 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 20
## step size given to GGDescent/Nesterov: 20
## objective: 44.28764 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 4
## step size given to GGDescent/Nesterov: 4
## objective: 44.22638 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 0.8
## step size given to GGDescent/Nesterov: 0.8
## objective: 44.21871 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 0.16
## step size given to GGDescent/Nesterov: 0.16
## objective: 44.2171 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 0.032
## step size given to GGDescent/Nesterov: 0.032
## objective: 44.21666 ( 200 iterations, max step size: 0.0064 )
## step size given to GGDescent/Nesterov: 0.032
## objective: 44.21652 ( 200 iterations, max step size: 0.0064 )
## step size given to GGDescent/Nesterov: 0.032
## objective: 44.21647 ( 200 iterations, max step size: 0.0064 )
## MM converged in 8 steps!
spcor_mat <- cov2cor(mm$Sigma)
#devtools::install_github("kkdey/CorShrink")
#library(CorShrink)
#sessionInfo()
cov_mat <- cov(data);
system.time(strimmer_sample <- corpcor::cov.shrink(data))
## Estimating optimal shrinkage intensity lambda.var (variance vector): 0.8871
##
## Estimating optimal shrinkage intensity lambda (correlation matrix): 0.8178
## user system elapsed
## 0.005 0.000 0.004
system.time(glasso_sample_005 <- glasso::glasso(cov_mat, rho = 0.05))
## user system elapsed
## 0.163 0.000 0.163
system.time(glasso_sample_05 <- glasso::glasso(cov_mat, rho = 0.5))
## user system elapsed
## 0.002 0.000 0.001
system.time(glasso_sample_1 <- glasso::glasso(cov_mat, rho = 1))
## user system elapsed
## 0.001 0.000 0.002
system.time(glasso_sample_10 <- glasso::glasso(cov_mat, rho = 10))
## user system elapsed
## 0.001 0.000 0.001
system.time(cov_sample_ML <- CorShrinkMatrix(cov2cor(cov_mat), nsamp = matrix(n, P, P), ash.control = list(mixcompdist = "normal", nullweight = 10)))
## ash cor only and ash cor PD matrices are different
## user system elapsed
## 3.604 0.095 3.712
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
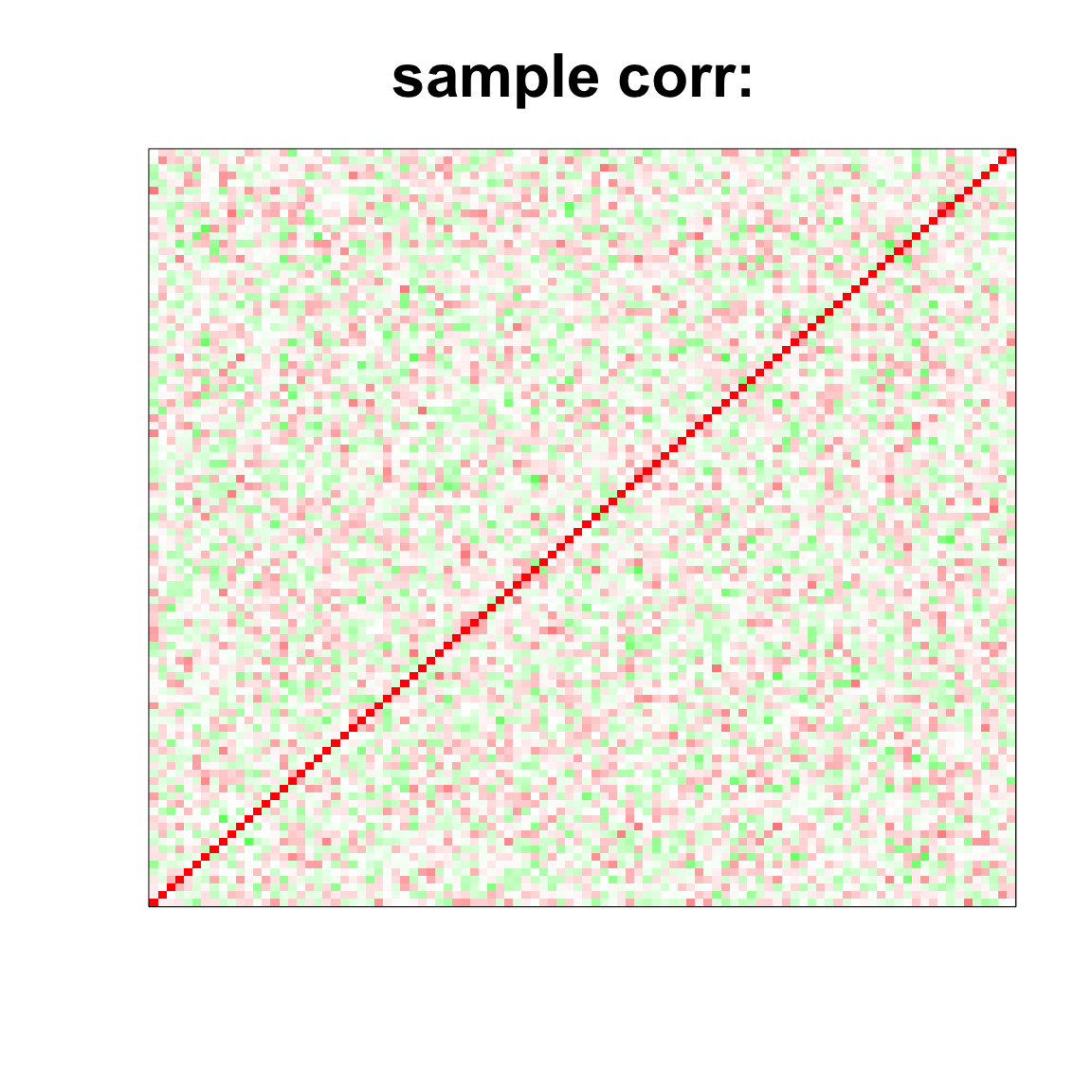
image(as.matrix(cov2cor(cov_mat)),
col=col, main=paste0("sample corr: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
par(mfrow = c(2,2))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(strimmer_sample)),
col=col, main=paste0("shafer strimmer: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov_sample_ML$ash_cor_only),
col=col, main=paste0("corshrink: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(glasso_sample_005$w)),
col=col, main=paste0("glasso 0.05: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(glasso_sample_05$w)),
col=col, main=paste0("glasso 0.5: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
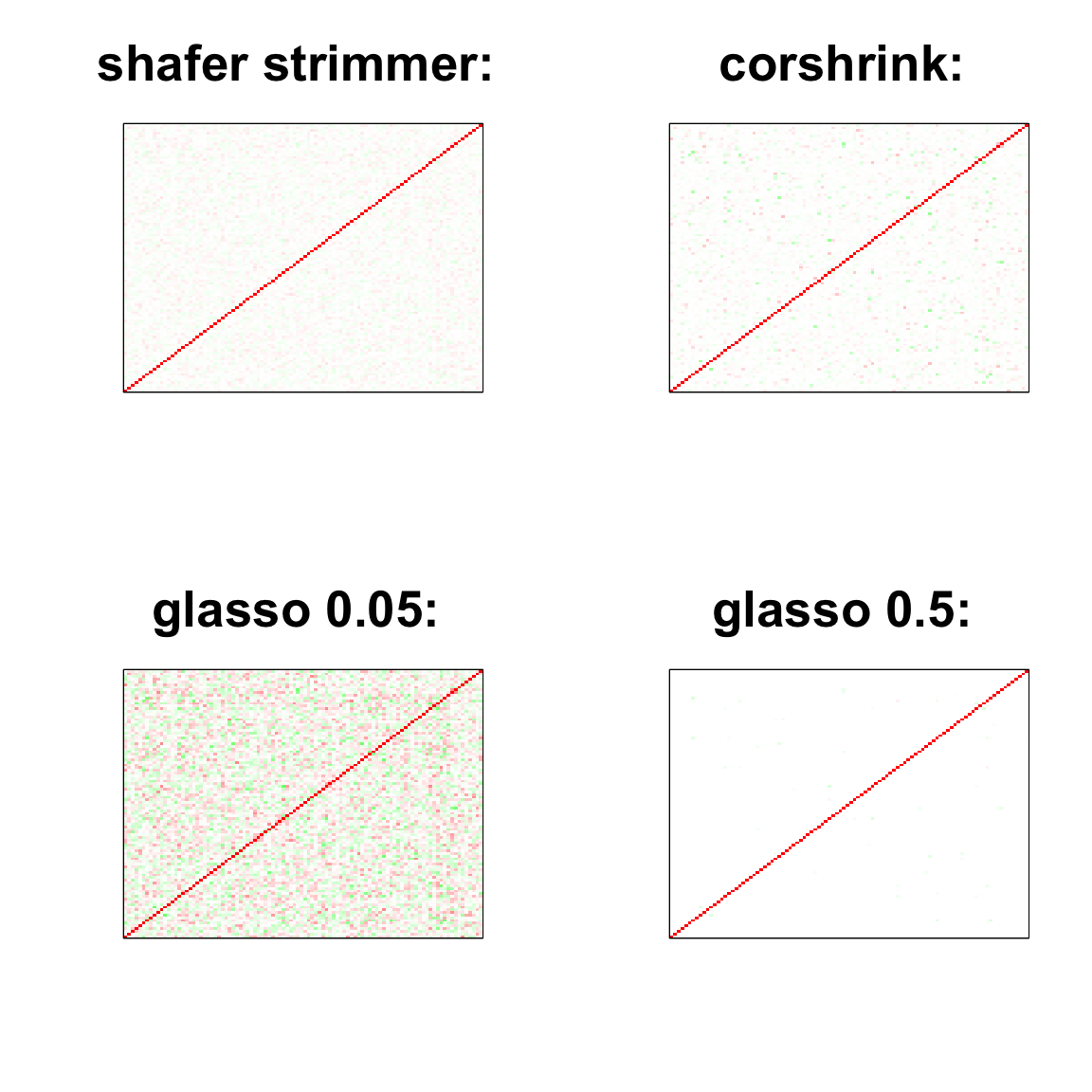
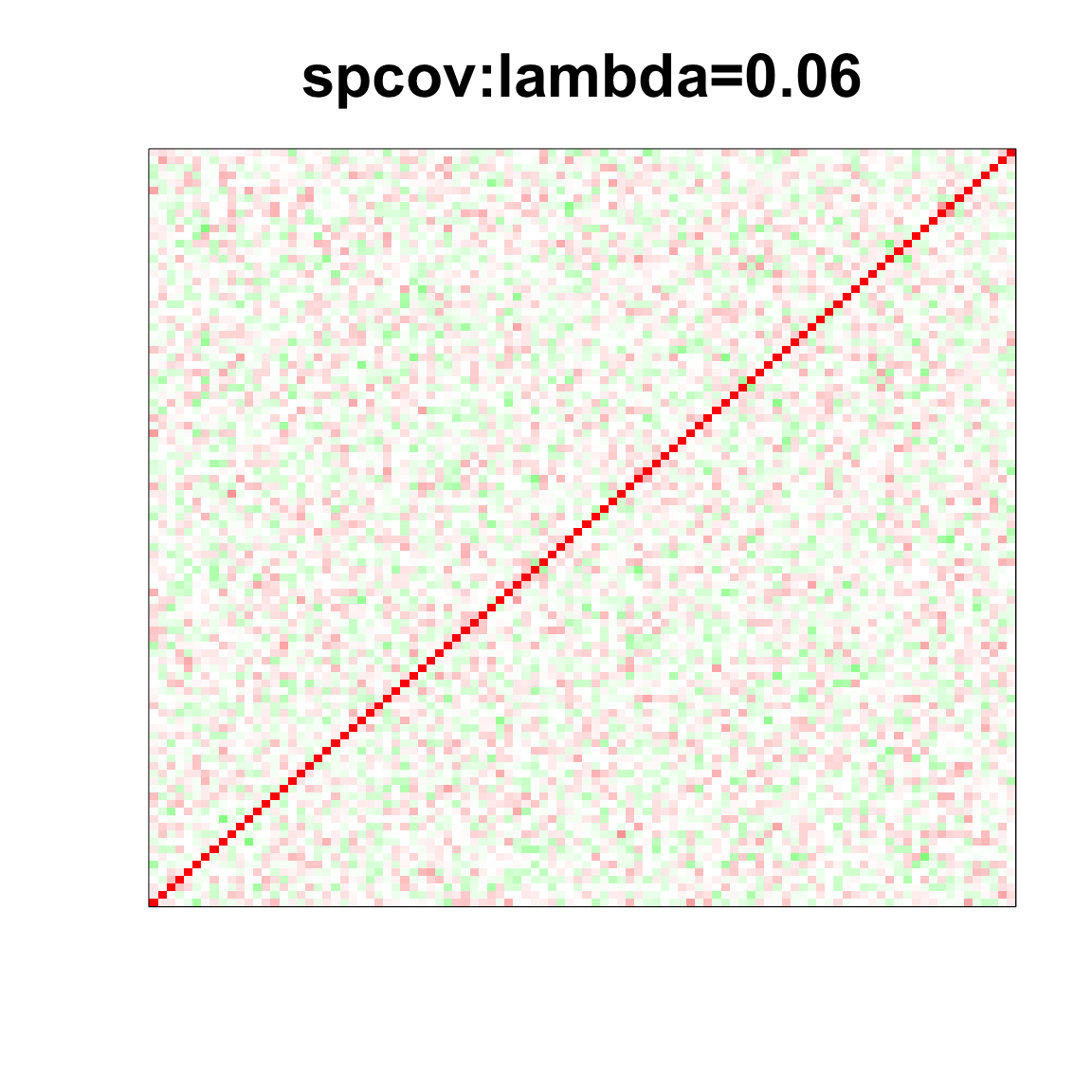
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(spcor_mat),
col=col, main=paste0("spcov:", expression(lambda), "=", lam), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
n = 5, P=100
n <- 5
P <- 100
ll <- DM_graph(n,P)
## Generating data from the multivariate normal distribution with the random graph structure....
## done.
data <- rbind(ll$Xtrain, ll$Xtest)
Sigma <- ll$Sigma
corSigma <- cov2cor(Sigma)
Pat <- matrix(1, P, P)
diag(Pat) <- 0
lam <- 0.06
step.size <- 100
tol <- 1e-4
covmat <- cov(data)
mm <- spcov(Sigma=covmat+0.1*diag(1,P), S=covmat+0.1*diag(1,P), lambda=lam * Pat,step.size=step.size, n.inner.steps=200, thr.inner=0, tol.outer=tol, trace=1)
## ---
## using Nesterov, backtracking line search
## ---
## objective: 113.7445
## step size given to GGDescent/Nesterov: 100
## objective: -59.55925 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 20
## step size given to GGDescent/Nesterov: 20
## objective: -62.38439 ( 200 iterations, max step size: 0.032 )
## Reducing step size to 4
## step size given to GGDescent/Nesterov: 4
## objective: -62.40129 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.8
## step size given to GGDescent/Nesterov: 0.8
## objective: -62.40288 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.16
## step size given to GGDescent/Nesterov: 0.16
## objective: -62.40329 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.032
## step size given to GGDescent/Nesterov: 0.032
## objective: -62.4034 ( 200 iterations, max step size: 0.00128 )
## step size given to GGDescent/Nesterov: 0.032
## objective: -62.40343 ( 200 iterations, max step size: 0.00128 )
## MM converged in 7 steps!
spcor_mat <- cov2cor(mm$Sigma)
#devtools::install_github("kkdey/CorShrink")
#library(CorShrink)
#sessionInfo()
cov_mat <- cov(data);
system.time(strimmer_sample <- corpcor::cov.shrink(data))
## Estimating optimal shrinkage intensity lambda.var (variance vector): 0.8824
##
## Estimating optimal shrinkage intensity lambda (correlation matrix): 0.7539
## user system elapsed
## 0.002 0.000 0.001
system.time(glasso_sample_005 <- glasso::glasso(cov_mat, rho = 0.05))
## user system elapsed
## 0.666 0.002 0.673
system.time(glasso_sample_05 <- glasso::glasso(cov_mat, rho = 0.5))
## user system elapsed
## 0.057 0.000 0.056
system.time(glasso_sample_1 <- glasso::glasso(cov_mat, rho = 1))
## user system elapsed
## 0.009 0.000 0.009
system.time(glasso_sample_10 <- glasso::glasso(cov_mat, rho = 10))
## user system elapsed
## 0.001 0.000 0.001
system.time(cov_sample_ML <- CorShrinkMatrix(cov2cor(cov_mat), nsamp = matrix(n, P, P), ash.control = list(mixcompdist = "normal", nullweight = 10)))
## ash cor only and ash cor PD matrices are different
## user system elapsed
## 1.860 0.048 1.914
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
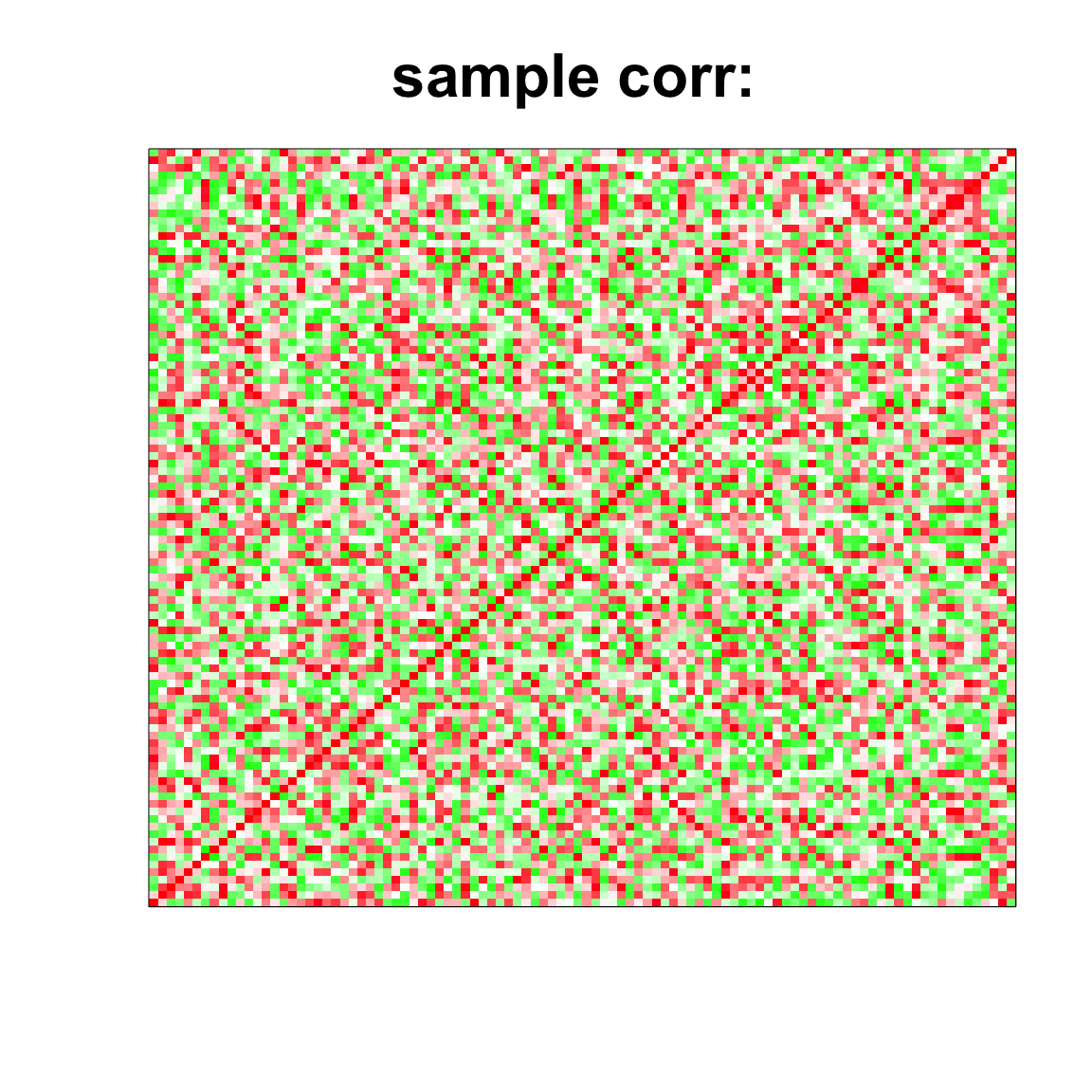
image(as.matrix(cov2cor(cov_mat)),
col=col, main=paste0("sample corr: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
par(mfrow = c(2,2))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
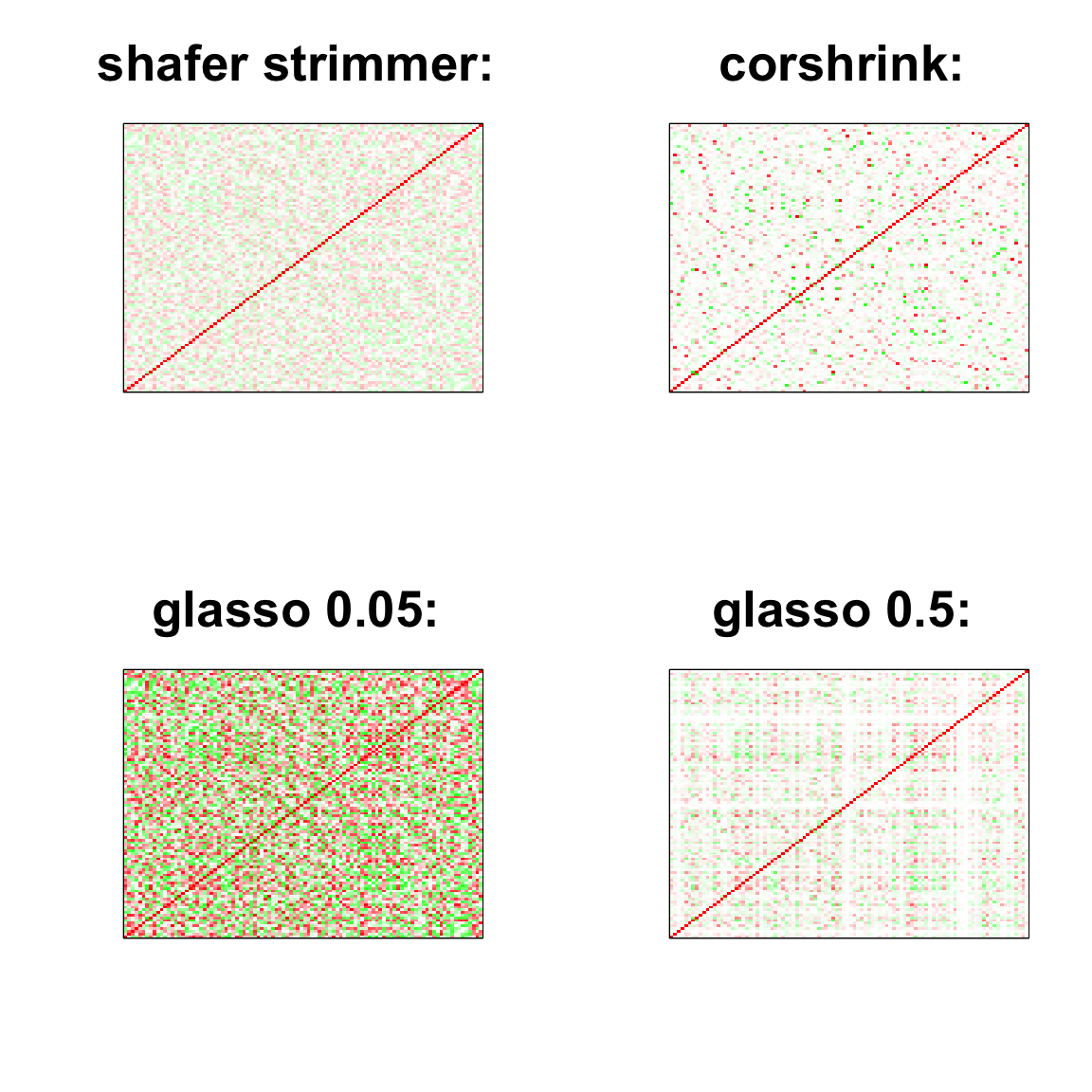
image(as.matrix(cov2cor(strimmer_sample)),
col=col, main=paste0("shafer strimmer: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov_sample_ML$ash_cor_only),
col=col, main=paste0("corshrink: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(glasso_sample_005$w)),
col=col, main=paste0("glasso 0.05: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(glasso_sample_05$w)),
col=col, main=paste0("glasso 0.5: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
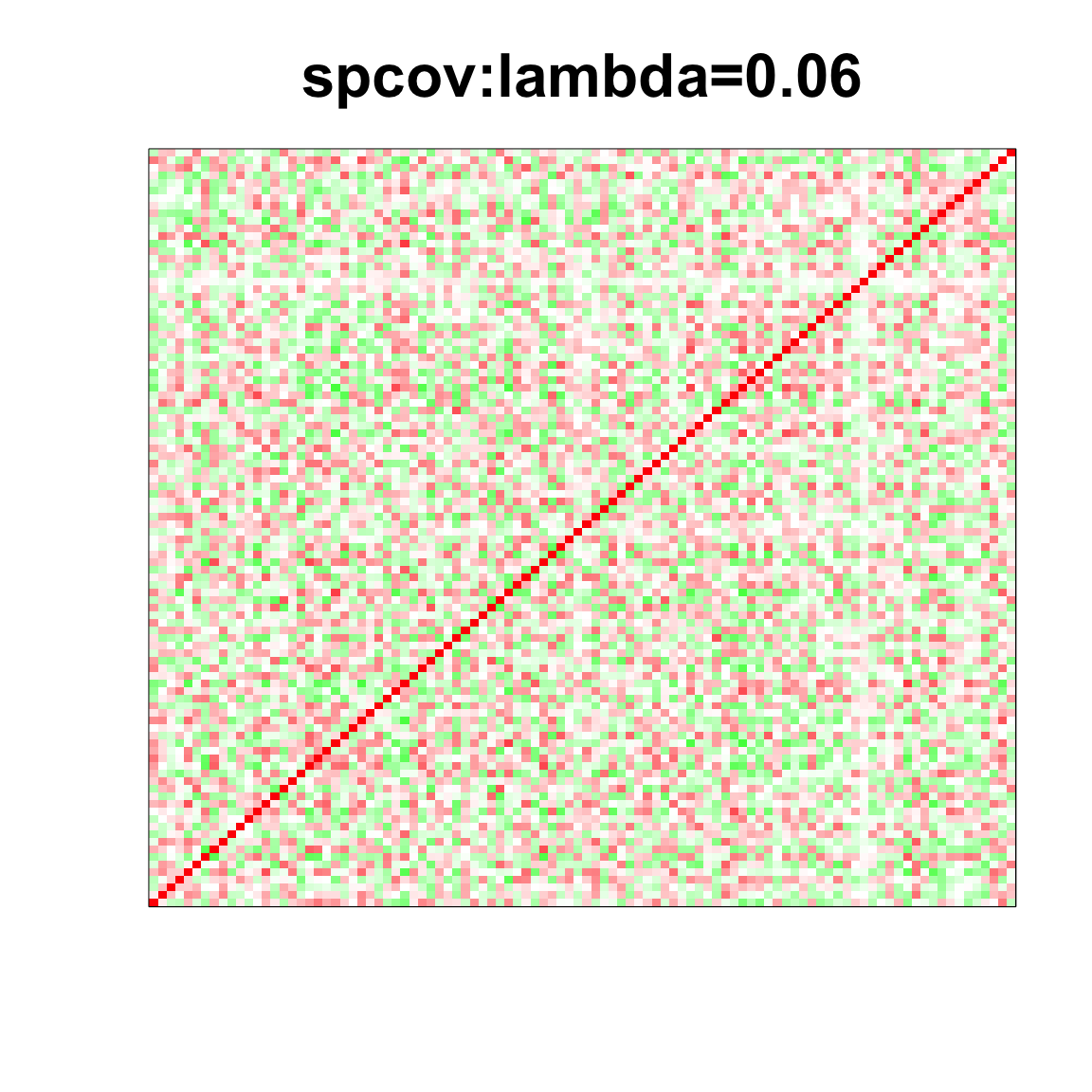
image(as.matrix(spcor_mat),
col=col, main=paste0("spcov:", expression(lambda), "=", lam), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
DM graph (hub graph)
n = 10, P=100
n <- 10
P <- 100
ll <- DM_graph(n,P,g_type = "hub", para = 5)
## Generating data from the multivariate normal distribution with the hub graph structure....
## done.
data <- rbind(ll$Xtrain, ll$Xtest)
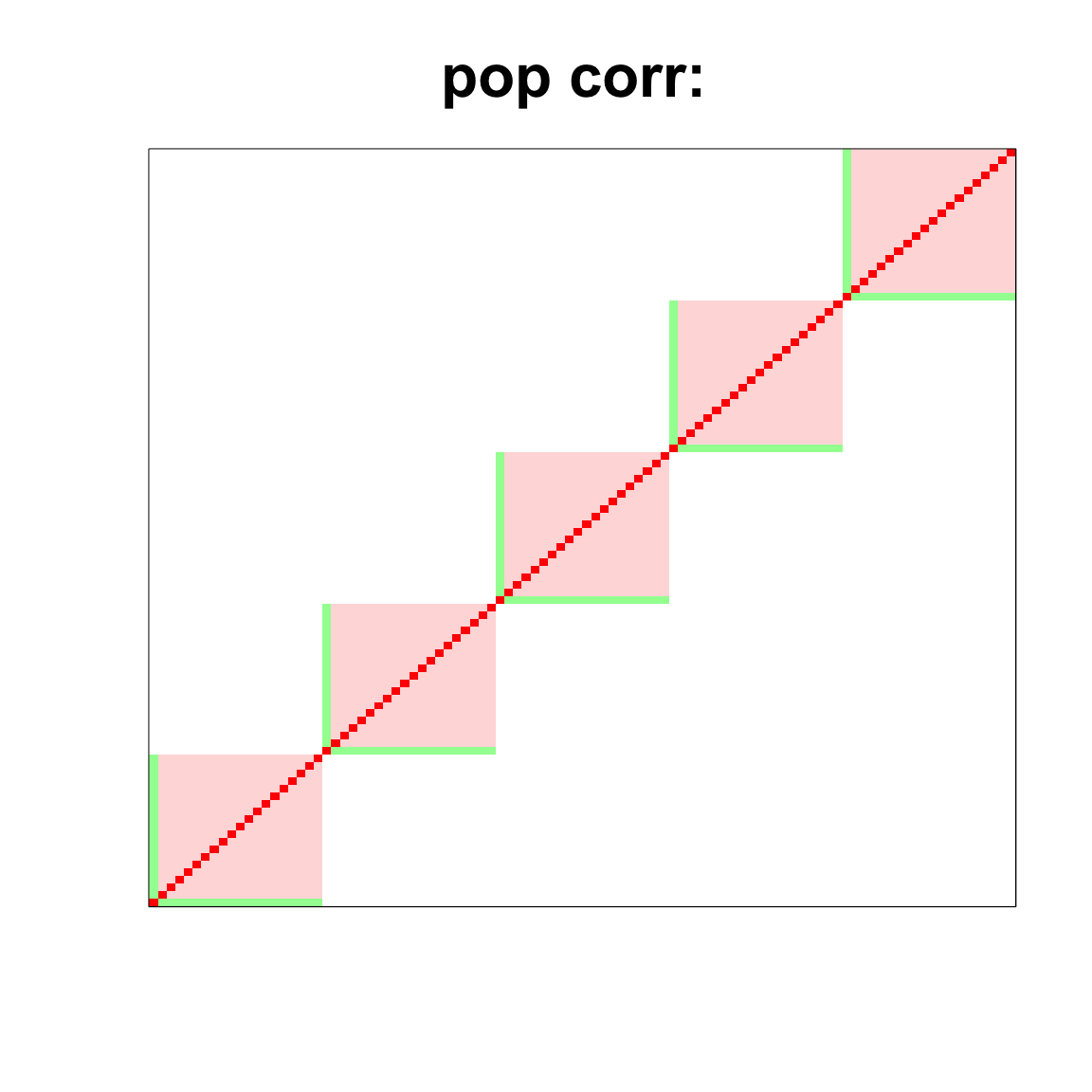
Sigma <- ll$Sigma
corSigma <- cov2cor(Sigma)
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(corSigma),
col=col, main=paste0("pop corr: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
Pat <- matrix(1, P, P)
diag(Pat) <- 0
lam <- 0.06
step.size <- 100
tol <- 1e-4
covmat <- cov(data)
mm <- spcov(Sigma=covmat+0.1*diag(1,P), S=covmat+0.1*diag(1,P), lambda=lam * Pat,step.size=step.size, n.inner.steps=200, thr.inner=0, tol.outer=tol, trace=1)
## ---
## using Nesterov, backtracking line search
## ---
## objective: 72.53971
## step size given to GGDescent/Nesterov: 100
## objective: -37.13113 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 20
## step size given to GGDescent/Nesterov: 20
## objective: -38.34797 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 4
## step size given to GGDescent/Nesterov: 4
## objective: -38.36321 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.8
## step size given to GGDescent/Nesterov: 0.8
## objective: -38.3649 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.16
## step size given to GGDescent/Nesterov: 0.16
## objective: -38.3653 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.032
## step size given to GGDescent/Nesterov: 0.032
## objective: -38.36541 ( 200 iterations, max step size: 0.00128 )
## step size given to GGDescent/Nesterov: 0.032
## objective: -38.36544 ( 200 iterations, max step size: 0.00128 )
## MM converged in 7 steps!
spcor_mat <- cov2cor(mm$Sigma)
#devtools::install_github("kkdey/CorShrink")
#library(CorShrink)
#sessionInfo()
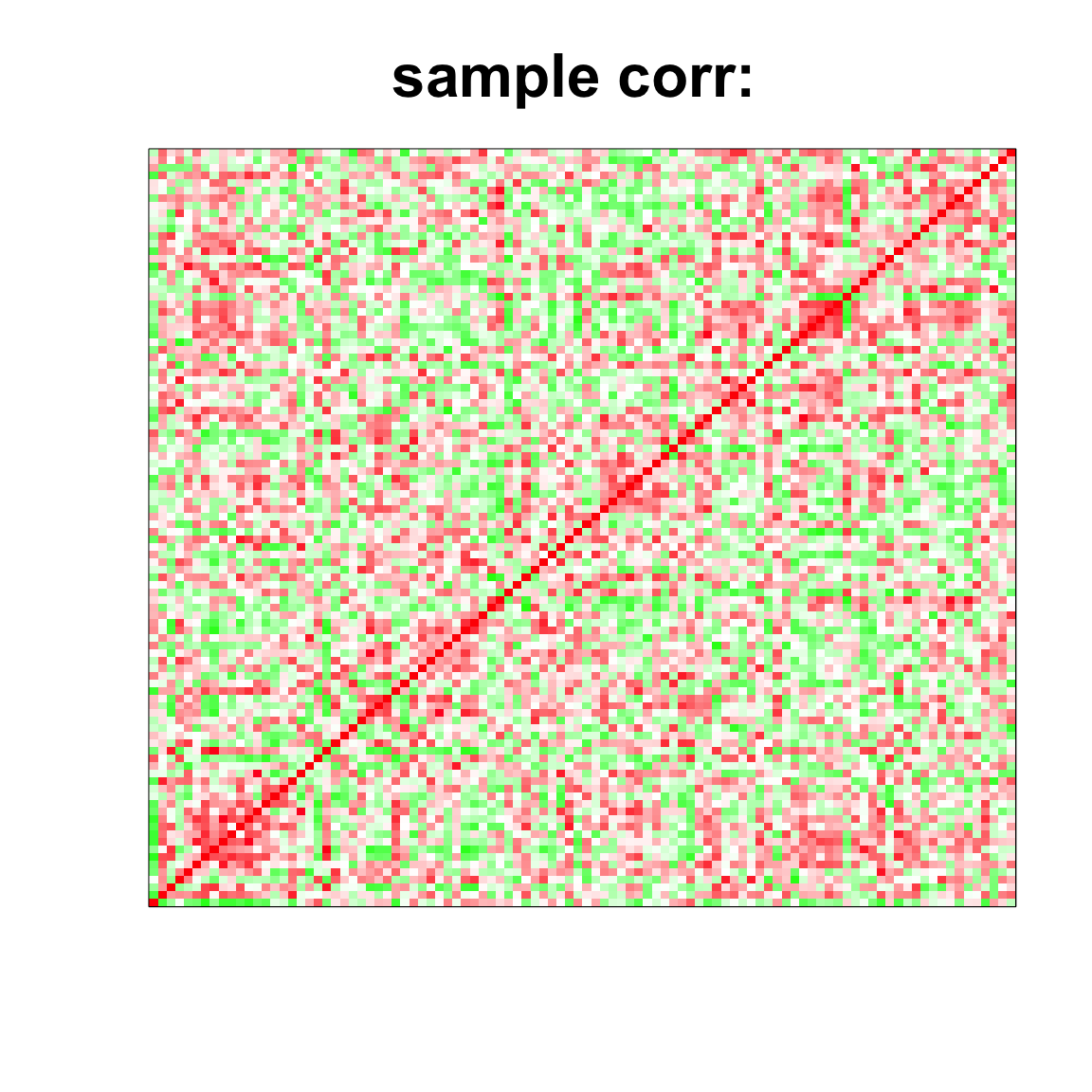
cov_mat <- cov(data);
system.time(strimmer_sample <- corpcor::cov.shrink(data))
## Estimating optimal shrinkage intensity lambda.var (variance vector): 0.982
##
## Estimating optimal shrinkage intensity lambda (correlation matrix): 0.8634
## user system elapsed
## 0.002 0.000 0.002
system.time(glasso_sample_005 <- glasso::glasso(cov_mat, rho = 0.05))
## user system elapsed
## 0.388 0.001 0.390
system.time(glasso_sample_05 <- glasso::glasso(cov_mat, rho = 0.5))
## user system elapsed
## 0.032 0.000 0.034
system.time(glasso_sample_1 <- glasso::glasso(cov_mat, rho = 1))
## user system elapsed
## 0.002 0.000 0.001
system.time(glasso_sample_10 <- glasso::glasso(cov_mat, rho = 10))
## user system elapsed
## 0.001 0.000 0.001
system.time(cov_sample_ML <- CorShrinkMatrix(cov2cor(cov_mat), nsamp = matrix(n, P, P), ash.control = list(mixcompdist = "normal", nullweight = 10)))
## ash cor only and ash cor PD matrices are different
## user system elapsed
## 1.965 0.050 2.053
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(cov_mat)),
col=col, main=paste0("sample corr: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
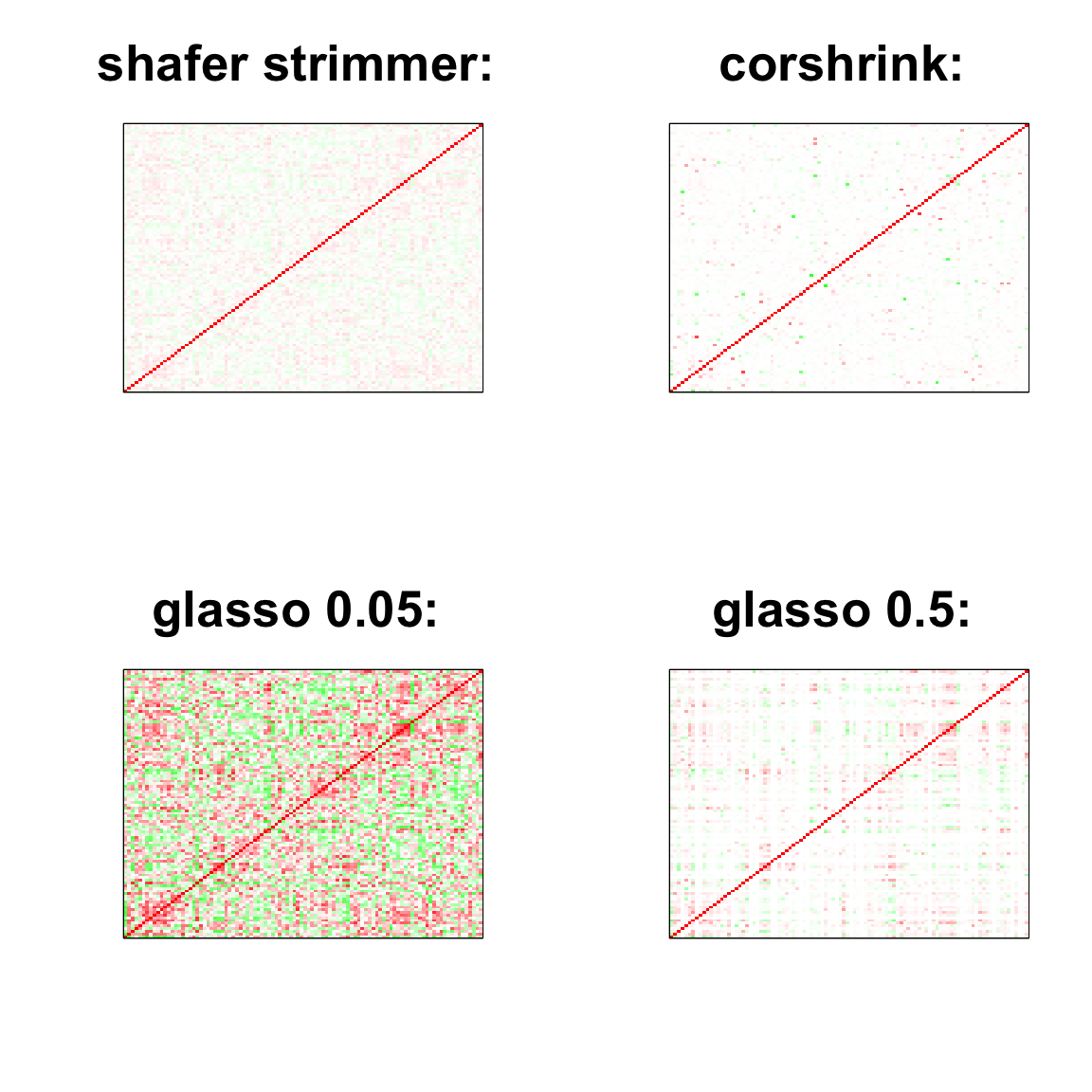
par(mfrow = c(2,2))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(strimmer_sample)),
col=col, main=paste0("shafer strimmer: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov_sample_ML$ash_cor_only),
col=col, main=paste0("corshrink: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(glasso_sample_005$w)),
col=col, main=paste0("glasso 0.05: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(glasso_sample_05$w)),
col=col, main=paste0("glasso 0.5: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
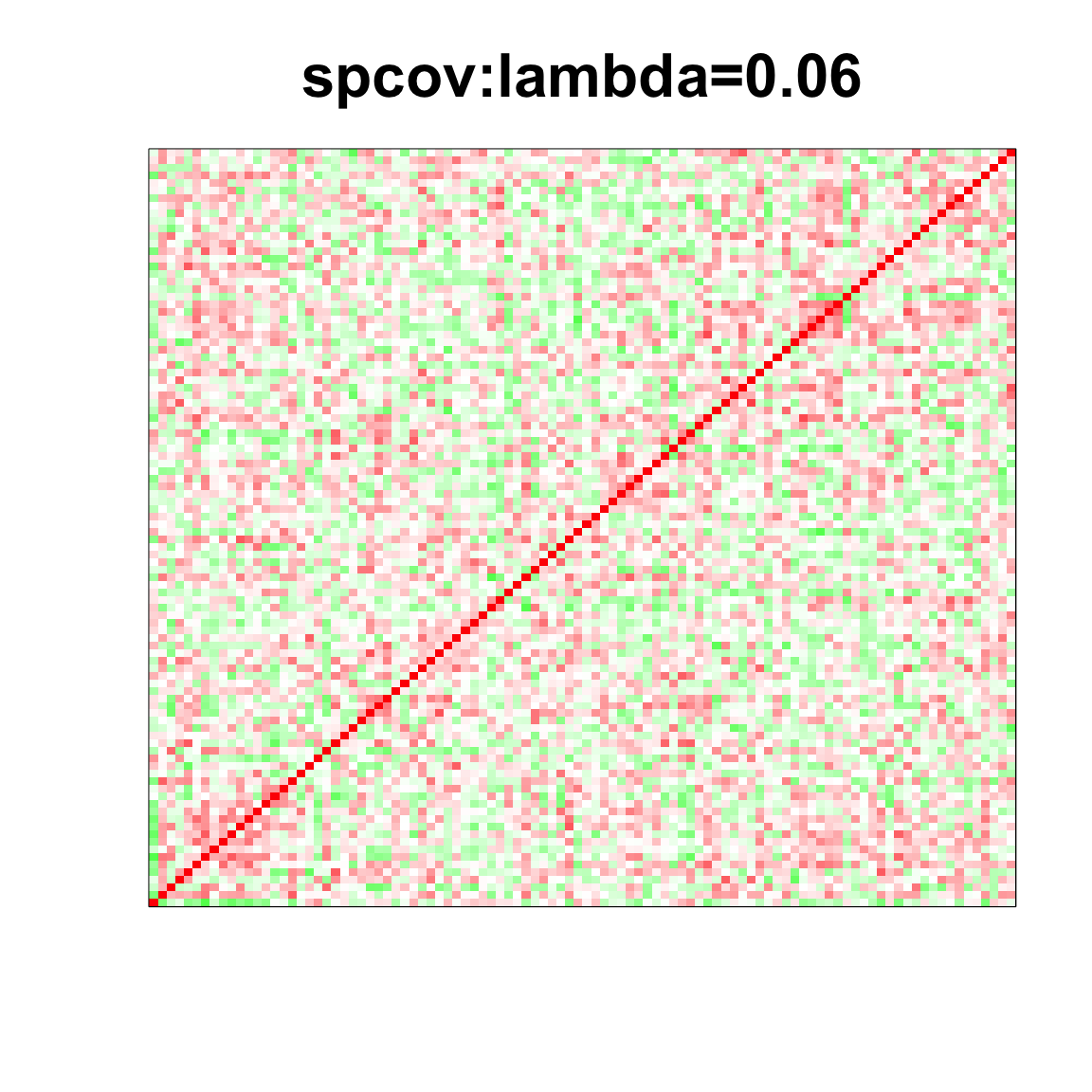
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(spcor_mat),
col=col, main=paste0("spcov:", expression(lambda), "=", lam), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
n = 50, P=100
n <- 50
P <- 100
ll <- DM_graph(n,P,g_type = "hub", para = 5)
## Generating data from the multivariate normal distribution with the hub graph structure....
## done.
data <- rbind(ll$Xtrain, ll$Xtest)
Sigma <- ll$Sigma
corSigma <- cov2cor(Sigma)
Pat <- matrix(1, P, P)
diag(Pat) <- 0
lam <- 0.06
step.size <- 100
tol <- 1e-4
covmat <- cov(data)
mm <- spcov(Sigma=covmat+0.1*diag(1,P), S=covmat+0.1*diag(1,P), lambda=lam * Pat,step.size=step.size, n.inner.steps=200, thr.inner=0, tol.outer=tol, trace=1)
## ---
## using Nesterov, backtracking line search
## ---
## objective: 77.0732
## step size given to GGDescent/Nesterov: 100
## objective: 40.91093 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 20
## step size given to GGDescent/Nesterov: 20
## objective: 40.05197 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 4
## step size given to GGDescent/Nesterov: 4
## objective: 39.99288 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 0.8
## step size given to GGDescent/Nesterov: 0.8
## objective: 39.98539 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 0.16
## step size given to GGDescent/Nesterov: 0.16
## objective: 39.98384 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 0.032
## step size given to GGDescent/Nesterov: 0.032
## objective: 39.98342 ( 200 iterations, max step size: 0.0064 )
## step size given to GGDescent/Nesterov: 0.032
## objective: 39.9833 ( 200 iterations, max step size: 0.0064 )
## step size given to GGDescent/Nesterov: 0.032
## objective: 39.98326 ( 200 iterations, max step size: 0.0064 )
## MM converged in 8 steps!
spcor_mat <- cov2cor(mm$Sigma)
#devtools::install_github("kkdey/CorShrink")
#library(CorShrink)
#sessionInfo()
cov_mat <- cov(data);
system.time(strimmer_sample <- corpcor::cov.shrink(data))
## Estimating optimal shrinkage intensity lambda.var (variance vector): 1
##
## Estimating optimal shrinkage intensity lambda (correlation matrix): 0.7481
## user system elapsed
## 0.005 0.000 0.005
system.time(glasso_sample_005 <- glasso::glasso(cov_mat, rho = 0.05))
## user system elapsed
## 0.167 0.001 0.172
system.time(glasso_sample_05 <- glasso::glasso(cov_mat, rho = 0.5))
## user system elapsed
## 0.001 0.000 0.001
system.time(glasso_sample_1 <- glasso::glasso(cov_mat, rho = 1))
## user system elapsed
## 0.003 0.000 0.004
system.time(glasso_sample_10 <- glasso::glasso(cov_mat, rho = 10))
## user system elapsed
## 0.001 0.000 0.001
system.time(cov_sample_ML <- CorShrinkMatrix(cov2cor(cov_mat), nsamp = matrix(n, P, P), ash.control = list(mixcompdist = "normal", nullweight = 10)))
## ash cor only and ash cor PD matrices are different
## user system elapsed
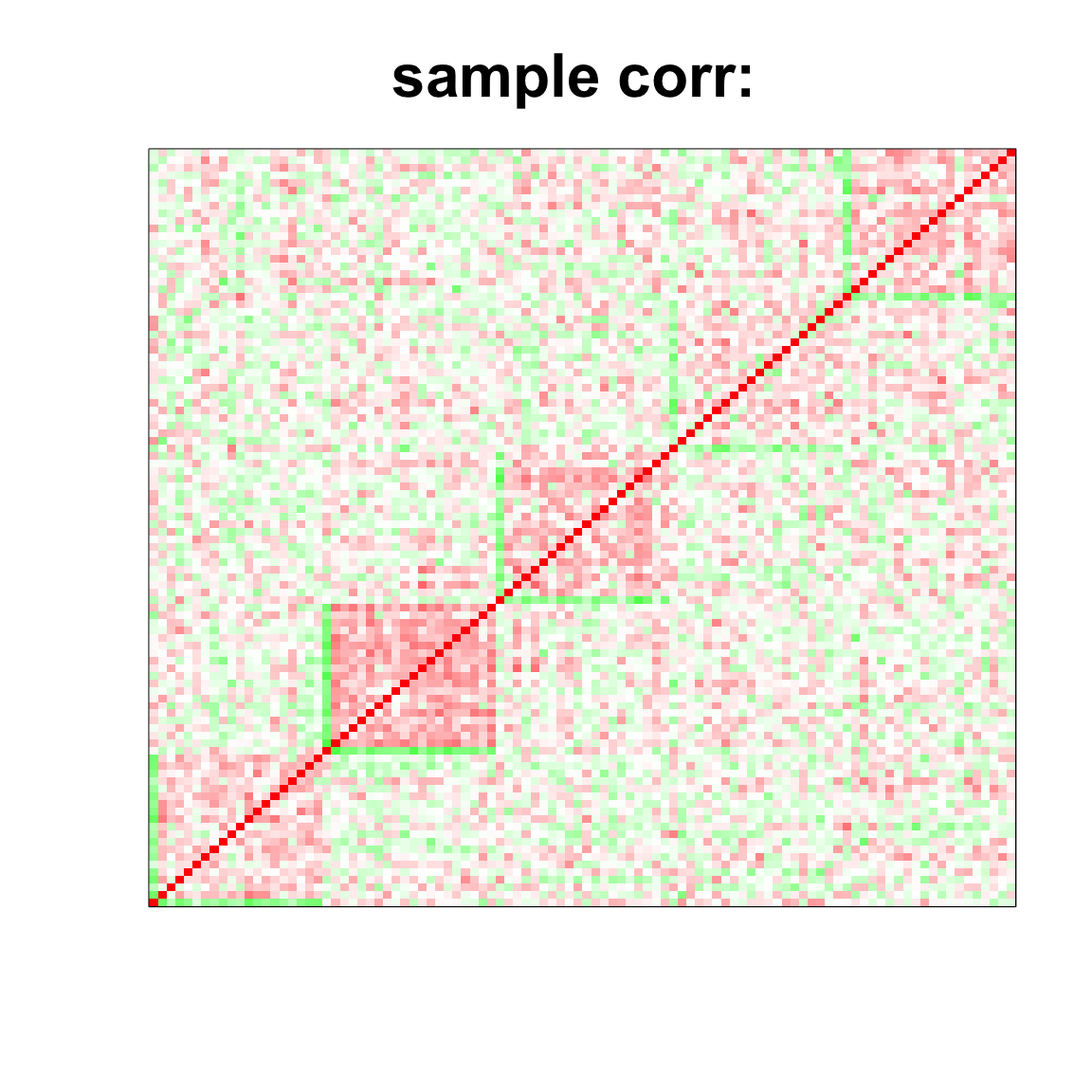
## 6.101 0.174 6.458
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(cov_mat)),
col=col, main=paste0("sample corr: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
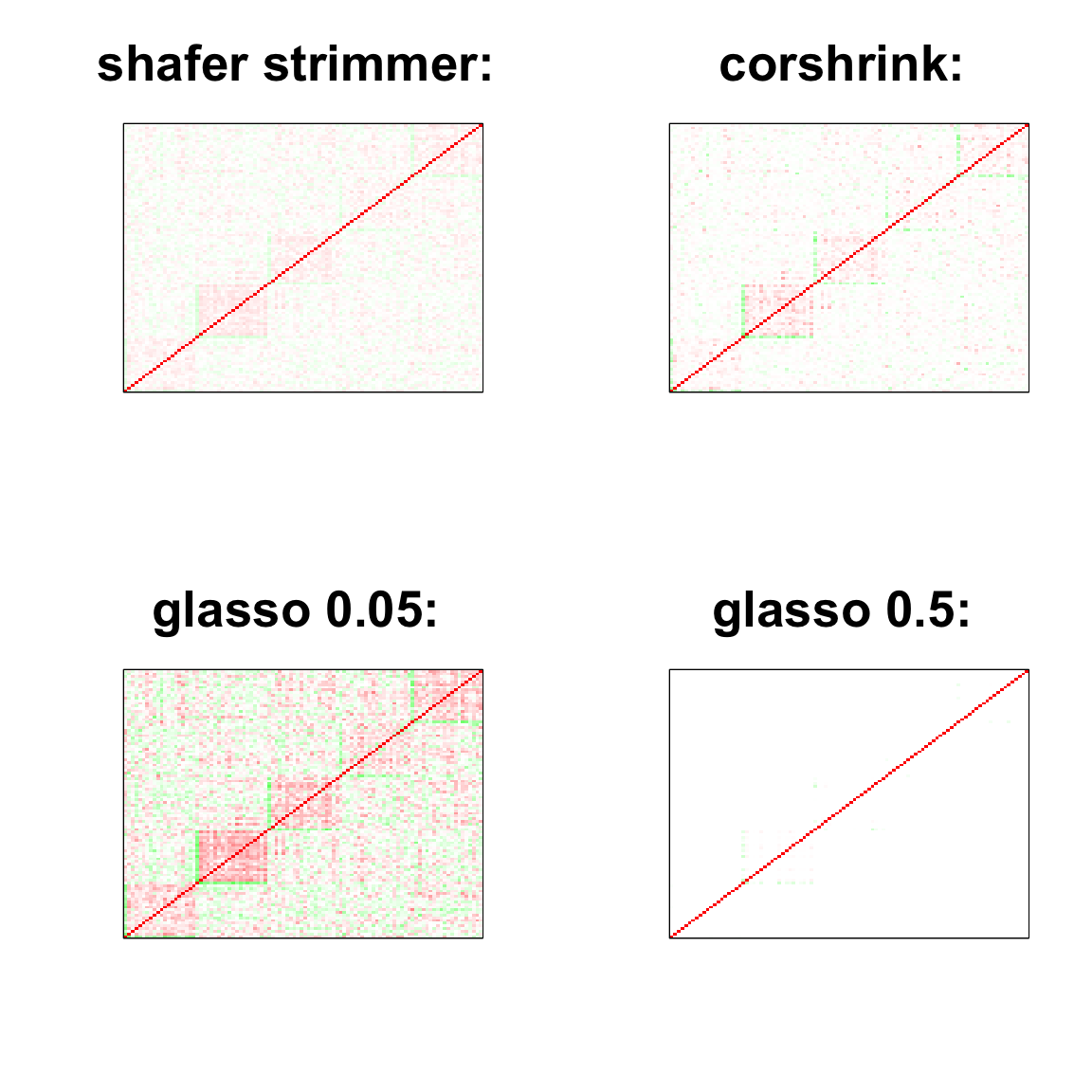
par(mfrow = c(2,2))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(strimmer_sample)),
col=col, main=paste0("shafer strimmer: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov_sample_ML$ash_cor_only),
col=col, main=paste0("corshrink: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(glasso_sample_005$w)),
col=col, main=paste0("glasso 0.05: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(glasso_sample_05$w)),
col=col, main=paste0("glasso 0.5: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
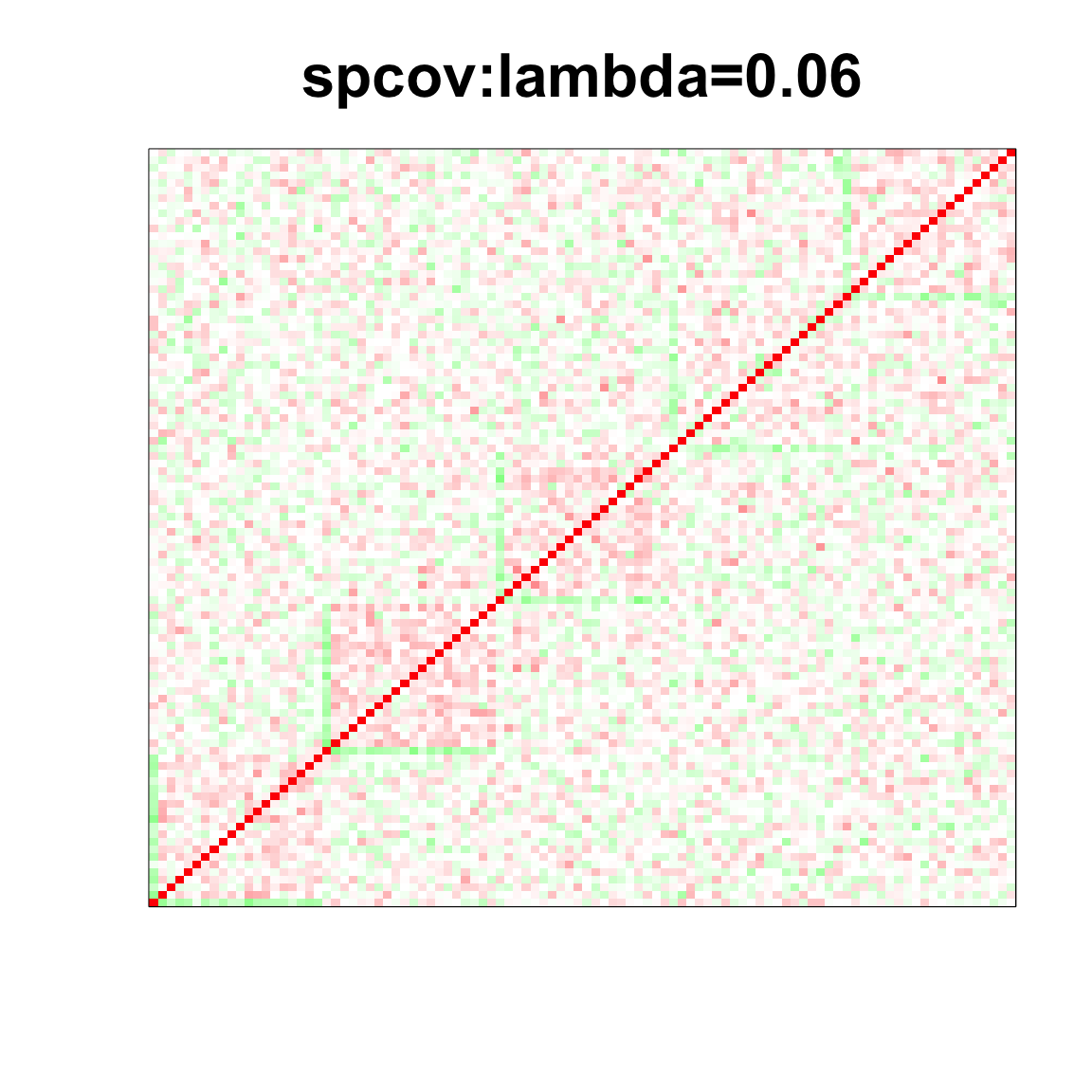
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(spcor_mat),
col=col, main=paste0("spcov:", expression(lambda), "=", lam), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
n = 5, P=100
n <- 5
P <- 100
ll <- DM_graph(n,P,g_type = "hub", para = 5)
## Generating data from the multivariate normal distribution with the hub graph structure....
## done.
data <- rbind(ll$Xtrain, ll$Xtest)
Sigma <- ll$Sigma
corSigma <- cov2cor(Sigma)
Pat <- matrix(1, P, P)
diag(Pat) <- 0
lam <- 0.06
step.size <- 100
tol <- 1e-4
covmat <- cov(data)
mm <- spcov(Sigma=covmat+0.1*diag(1,P), S=covmat+0.1*diag(1,P), lambda=lam * Pat,step.size=step.size, n.inner.steps=200, thr.inner=0, tol.outer=tol, trace=1)
## ---
## using Nesterov, backtracking line search
## ---
## objective: 95.50814
## step size given to GGDescent/Nesterov: 100
## objective: -62.61963 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 20
## step size given to GGDescent/Nesterov: 20
## objective: -65.07446 ( 200 iterations, max step size: 0.032 )
## Reducing step size to 4
## step size given to GGDescent/Nesterov: 4
## objective: -65.09053 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.8
## step size given to GGDescent/Nesterov: 0.8
## objective: -65.09216 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.16
## step size given to GGDescent/Nesterov: 0.16
## objective: -65.09257 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.032
## step size given to GGDescent/Nesterov: 0.032
## objective: -65.09269 ( 200 iterations, max step size: 0.00128 )
## step size given to GGDescent/Nesterov: 0.032
## objective: -65.09272 ( 200 iterations, max step size: 0.00128 )
## MM converged in 7 steps!
spcor_mat <- cov2cor(mm$Sigma)
#devtools::install_github("kkdey/CorShrink")
#library(CorShrink)
#sessionInfo()
cov_mat <- cov(data);
system.time(strimmer_sample <- corpcor::cov.shrink(data))
## Estimating optimal shrinkage intensity lambda.var (variance vector): 0.7877
##
## Estimating optimal shrinkage intensity lambda (correlation matrix): 0.7607
## user system elapsed
## 0.002 0.000 0.002
system.time(glasso_sample_005 <- glasso::glasso(cov_mat, rho = 0.05))
## user system elapsed
## 0.572 0.004 0.583
system.time(glasso_sample_05 <- glasso::glasso(cov_mat, rho = 0.5))
## user system elapsed
## 0.054 0.000 0.054
system.time(glasso_sample_1 <- glasso::glasso(cov_mat, rho = 1))
## user system elapsed
## 0.007 0.000 0.007
system.time(glasso_sample_10 <- glasso::glasso(cov_mat, rho = 10))
## user system elapsed
## 0.002 0.000 0.001
system.time(cov_sample_ML <- CorShrinkMatrix(cov2cor(cov_mat), nsamp = matrix(n, P, P), ash.control = list(mixcompdist = "normal", nullweight = 10)))
## ash cor only and ash cor PD matrices are different
## user system elapsed
## 1.801 0.050 1.888
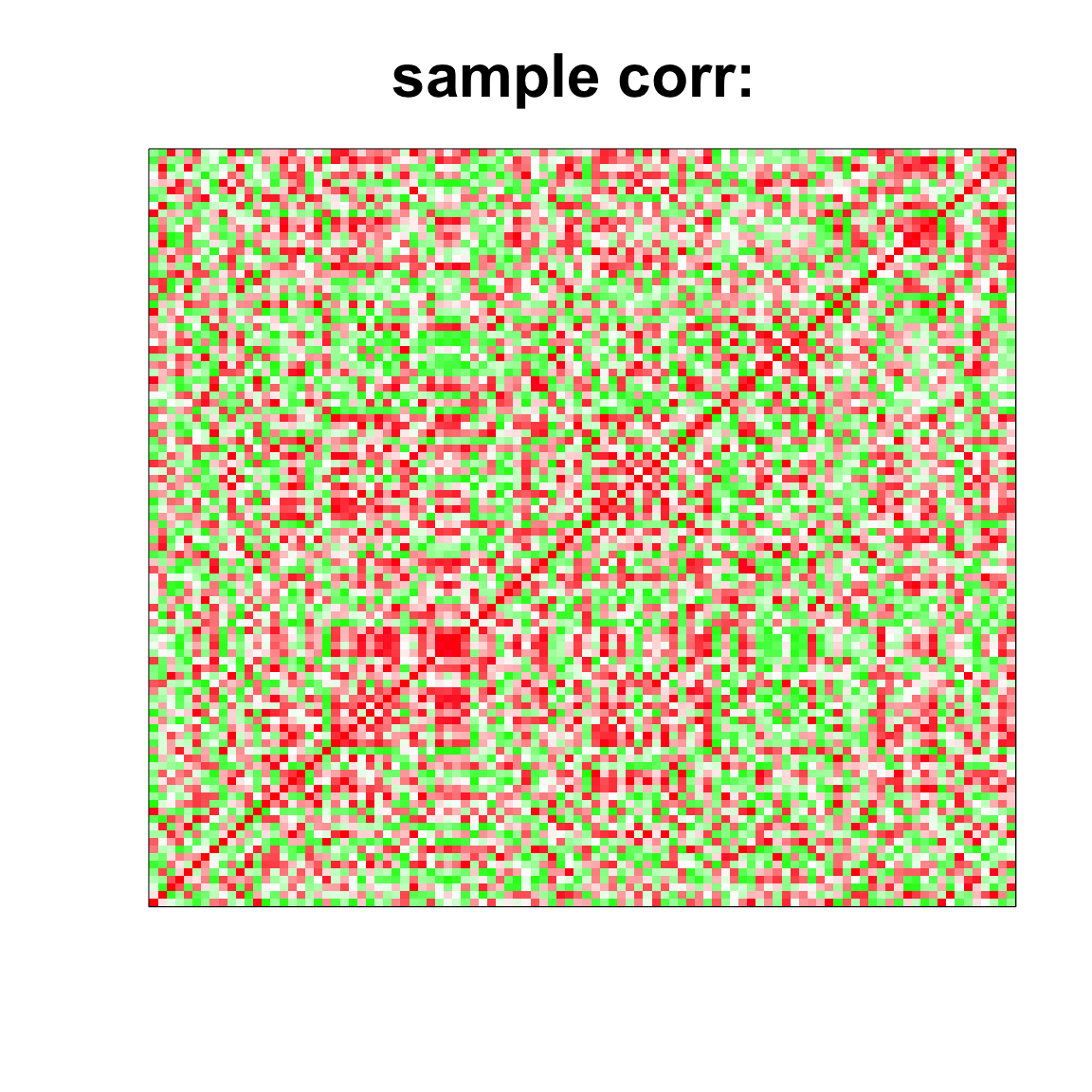
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(cov_mat)),
col=col, main=paste0("sample corr: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
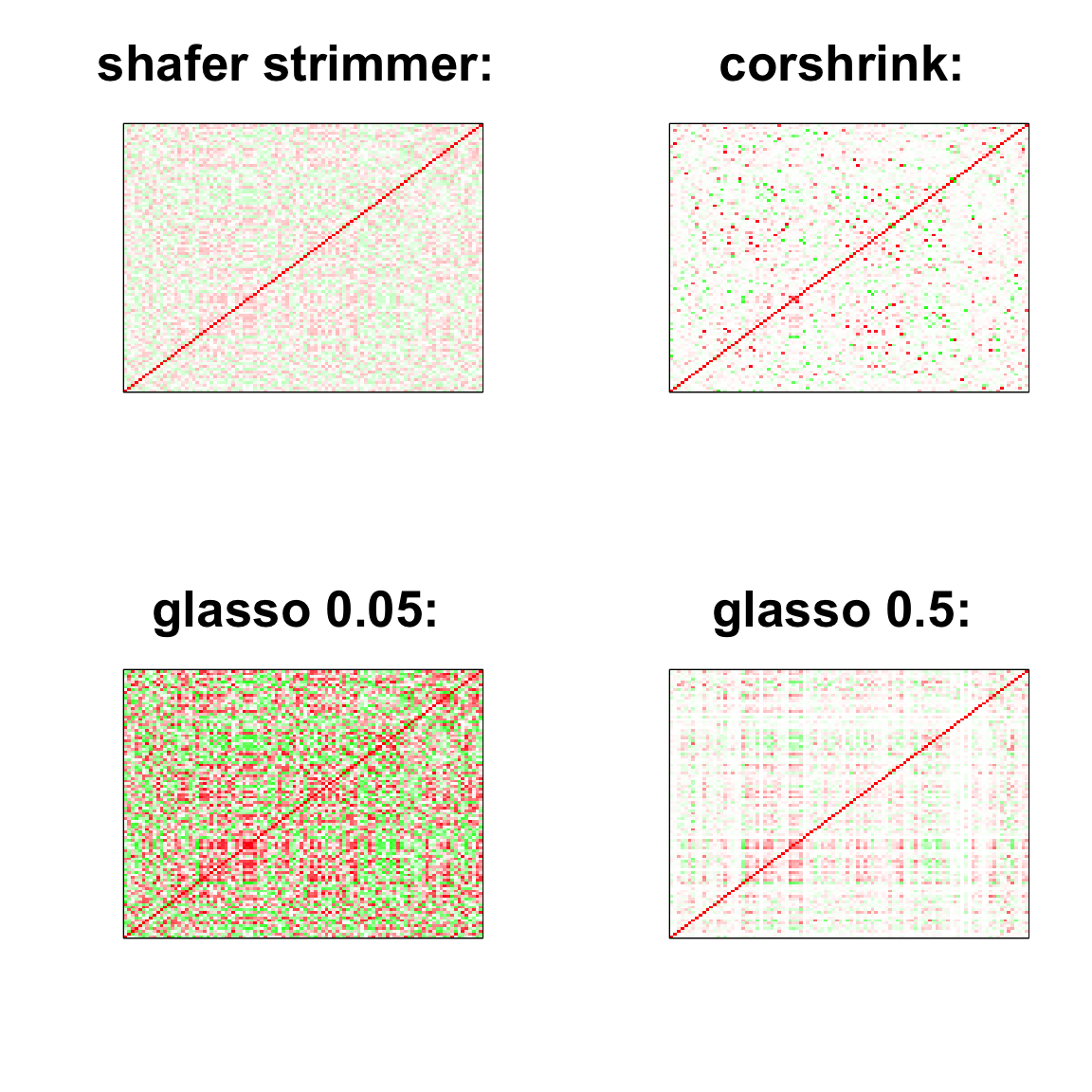
par(mfrow = c(2,2))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(strimmer_sample)),
col=col, main=paste0("shafer strimmer: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov_sample_ML$ash_cor_only),
col=col, main=paste0("corshrink: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(glasso_sample_005$w)),
col=col, main=paste0("glasso 0.05: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(glasso_sample_05$w)),
col=col, main=paste0("glasso 0.5: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
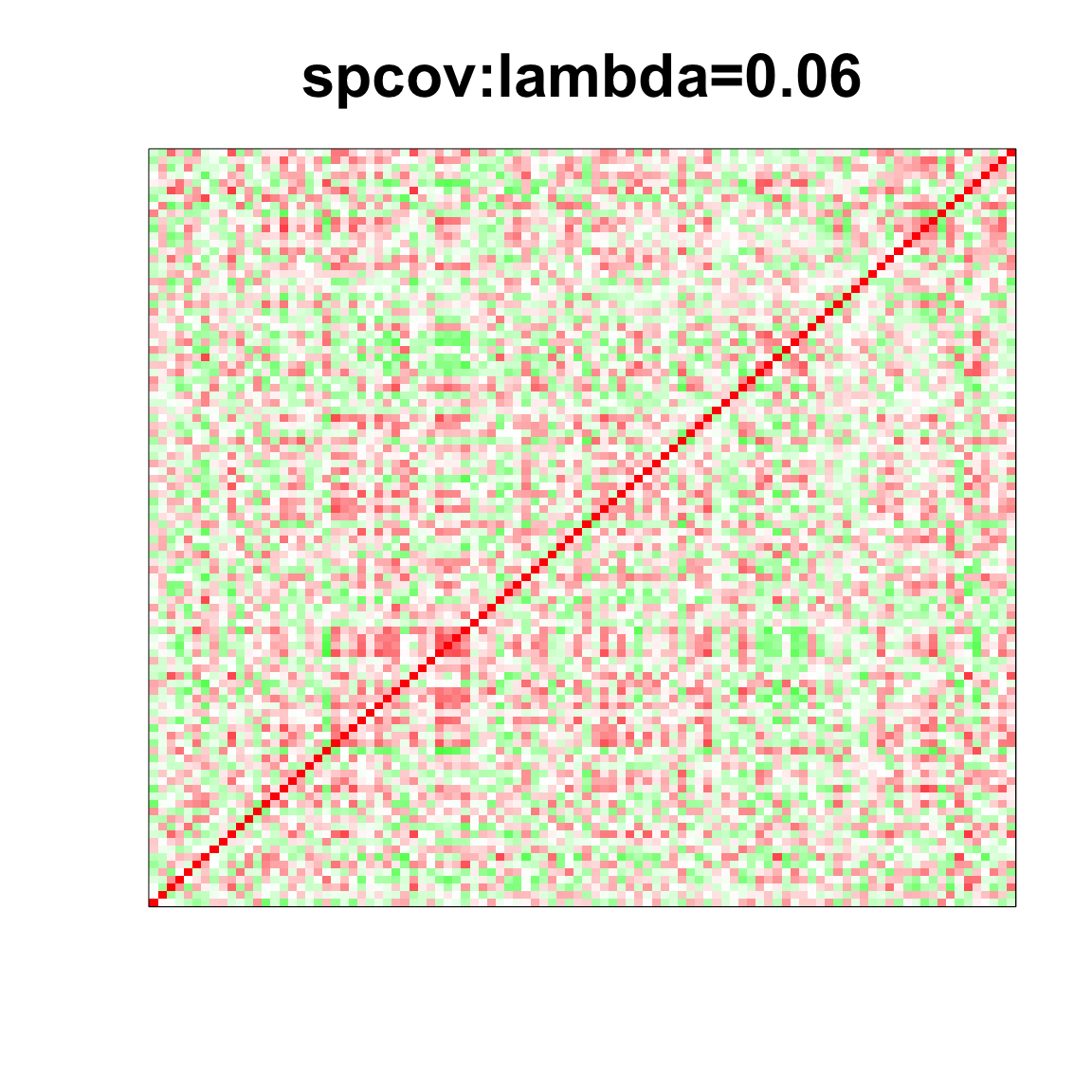
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(spcor_mat),
col=col, main=paste0("spcov:", expression(lambda), "=", lam), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
DM graph (scale free graph)
n = 10, P=100
n <- 10
P <- 100
ll <- DM_graph(n,P,g_type = "scale-free")
## Generating data from the multivariate normal distribution with the scale-free graph structure....
## done.
data <- rbind(ll$Xtrain, ll$Xtest)
Sigma <- ll$Sigma
corSigma <- cov2cor(Sigma)
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(corSigma),
col=col, main=paste0("pop corr: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
Pat <- matrix(1, P, P)
diag(Pat) <- 0
lam <- 0.06
step.size <- 100
tol <- 1e-4
covmat <- cov(data)
mm <- spcov(Sigma=covmat+0.1*diag(1,P), S=covmat+0.1*diag(1,P), lambda=lam * Pat,step.size=step.size, n.inner.steps=200, thr.inner=0, tol.outer=tol, trace=1)
## ---
## using Nesterov, backtracking line search
## ---
## objective: 71.9578
## step size given to GGDescent/Nesterov: 100
## objective: -38.07004 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 20
## step size given to GGDescent/Nesterov: 20
## objective: -38.68548 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 4
## step size given to GGDescent/Nesterov: 4
## objective: -38.69511 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.8
## step size given to GGDescent/Nesterov: 0.8
## objective: -38.69662 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.16
## step size given to GGDescent/Nesterov: 0.16
## objective: -38.697 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.032
## step size given to GGDescent/Nesterov: 0.032
## objective: -38.6971 ( 200 iterations, max step size: 0.00128 )
## step size given to GGDescent/Nesterov: 0.032
## objective: -38.69713 ( 200 iterations, max step size: 0.00128 )
## MM converged in 7 steps!
spcor_mat <- cov2cor(mm$Sigma)
#devtools::install_github("kkdey/CorShrink")
#library(CorShrink)
#sessionInfo()
cov_mat <- cov(data);
system.time(strimmer_sample <- corpcor::cov.shrink(data))
## Estimating optimal shrinkage intensity lambda.var (variance vector): 0.6248
##
## Estimating optimal shrinkage intensity lambda (correlation matrix): 0.8519
## user system elapsed
## 0.003 0.000 0.003
system.time(glasso_sample_005 <- glasso::glasso(cov_mat, rho = 0.05))
## user system elapsed
## 0.427 0.003 0.444
system.time(glasso_sample_05 <- glasso::glasso(cov_mat, rho = 0.5))
## user system elapsed
## 0.032 0.000 0.033
system.time(glasso_sample_1 <- glasso::glasso(cov_mat, rho = 1))
## user system elapsed
## 0.003 0.000 0.003
system.time(glasso_sample_10 <- glasso::glasso(cov_mat, rho = 10))
## user system elapsed
## 0.002 0.000 0.002
system.time(cov_sample_ML <- CorShrinkMatrix(cov2cor(cov_mat), nsamp = matrix(n, P, P), ash.control = list(mixcompdist = "normal", nullweight = 10)))
## ash cor only and ash cor PD matrices are different
## user system elapsed
## 3.939 0.125 4.203
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(cov_mat)),
col=col, main=paste0("sample corr: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
par(mfrow = c(2,2))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(strimmer_sample)),
col=col, main=paste0("shafer strimmer: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov_sample_ML$ash_cor_only),
col=col, main=paste0("corshrink: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(glasso_sample_005$w)),
col=col, main=paste0("glasso 0.05: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(glasso_sample_05$w)),
col=col, main=paste0("glasso 0.5: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(spcor_mat),
col=col, main=paste0("spcov:", expression(lambda), "=", lam), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
n = 50, P=100
n <- 50
P <- 100
ll <- DM_graph(n,P,g_type = "scale-free")
## Generating data from the multivariate normal distribution with the scale-free graph structure....
## done.
data <- rbind(ll$Xtrain, ll$Xtest)
Sigma <- ll$Sigma
corSigma <- cov2cor(Sigma)
Pat <- matrix(1, P, P)
diag(Pat) <- 0
lam <- 0.06
step.size <- 100
tol <- 1e-4
covmat <- cov(data)
mm <- spcov(Sigma=covmat+0.1*diag(1,P), S=covmat+0.1*diag(1,P), lambda=lam * Pat,step.size=step.size, n.inner.steps=200, thr.inner=0, tol.outer=tol, trace=1)
## ---
## using Nesterov, backtracking line search
## ---
## objective: 73.59023
## step size given to GGDescent/Nesterov: 100
## objective: 42.85039 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 20
## step size given to GGDescent/Nesterov: 20
## objective: 42.00787 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 4
## step size given to GGDescent/Nesterov: 4
## objective: 41.94759 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 0.8
## step size given to GGDescent/Nesterov: 0.8
## objective: 41.94014 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 0.16
## step size given to GGDescent/Nesterov: 0.16
## objective: 41.93864 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 0.032
## step size given to GGDescent/Nesterov: 0.032
## objective: 41.93824 ( 200 iterations, max step size: 0.0064 )
## step size given to GGDescent/Nesterov: 0.032
## objective: 41.93812 ( 200 iterations, max step size: 0.0064 )
## step size given to GGDescent/Nesterov: 0.032
## objective: 41.93808 ( 200 iterations, max step size: 0.0064 )
## MM converged in 8 steps!
spcor_mat <- cov2cor(mm$Sigma)
#devtools::install_github("kkdey/CorShrink")
#library(CorShrink)
#sessionInfo()
cov_mat <- cov(data);
system.time(strimmer_sample <- corpcor::cov.shrink(data))
## Estimating optimal shrinkage intensity lambda.var (variance vector): 0.986
##
## Estimating optimal shrinkage intensity lambda (correlation matrix): 0.8776
## user system elapsed
## 0.005 0.001 0.004
system.time(glasso_sample_005 <- glasso::glasso(cov_mat, rho = 0.05))
## user system elapsed
## 0.150 0.000 0.151
system.time(glasso_sample_05 <- glasso::glasso(cov_mat, rho = 0.5))
## user system elapsed
## 0.001 0.000 0.002
system.time(glasso_sample_1 <- glasso::glasso(cov_mat, rho = 1))
## user system elapsed
## 0.001 0.000 0.001
system.time(glasso_sample_10 <- glasso::glasso(cov_mat, rho = 10))
## user system elapsed
## 0.001 0.000 0.001
system.time(cov_sample_ML <- CorShrinkMatrix(cov2cor(cov_mat), nsamp = matrix(n, P, P), ash.control = list(mixcompdist = "normal", nullweight = 10)))
## ash cor only and ash cor PD matrices are different
## user system elapsed
## 2.724 0.059 2.793
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(cov_mat)),
col=col, main=paste0("sample corr: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
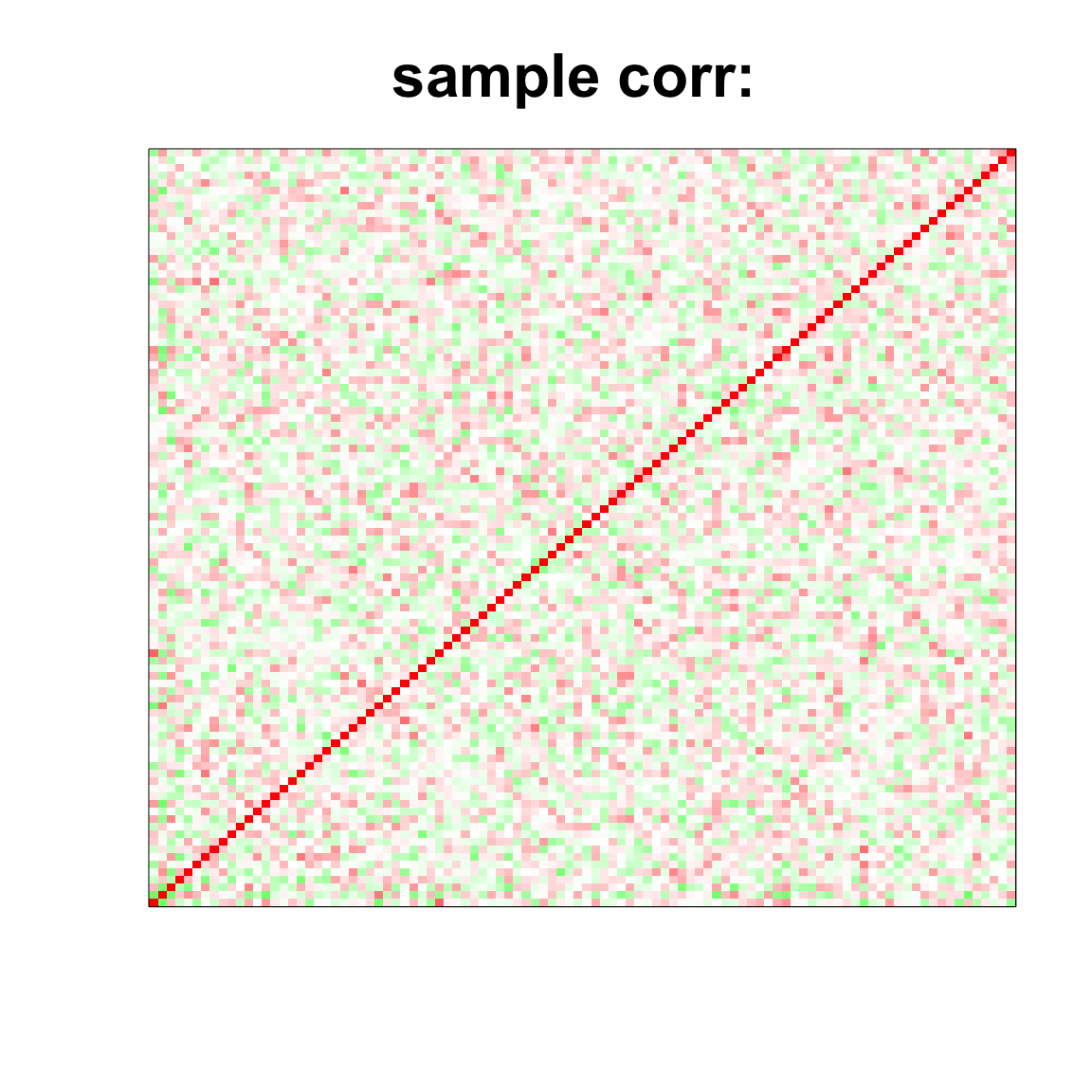
par(mfrow = c(2,2))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(strimmer_sample)),
col=col, main=paste0("shafer strimmer: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov_sample_ML$ash_cor_only),
col=col, main=paste0("corshrink: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(glasso_sample_005$w)),
col=col, main=paste0("glasso 0.05: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(glasso_sample_05$w)),
col=col, main=paste0("glasso 0.5: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
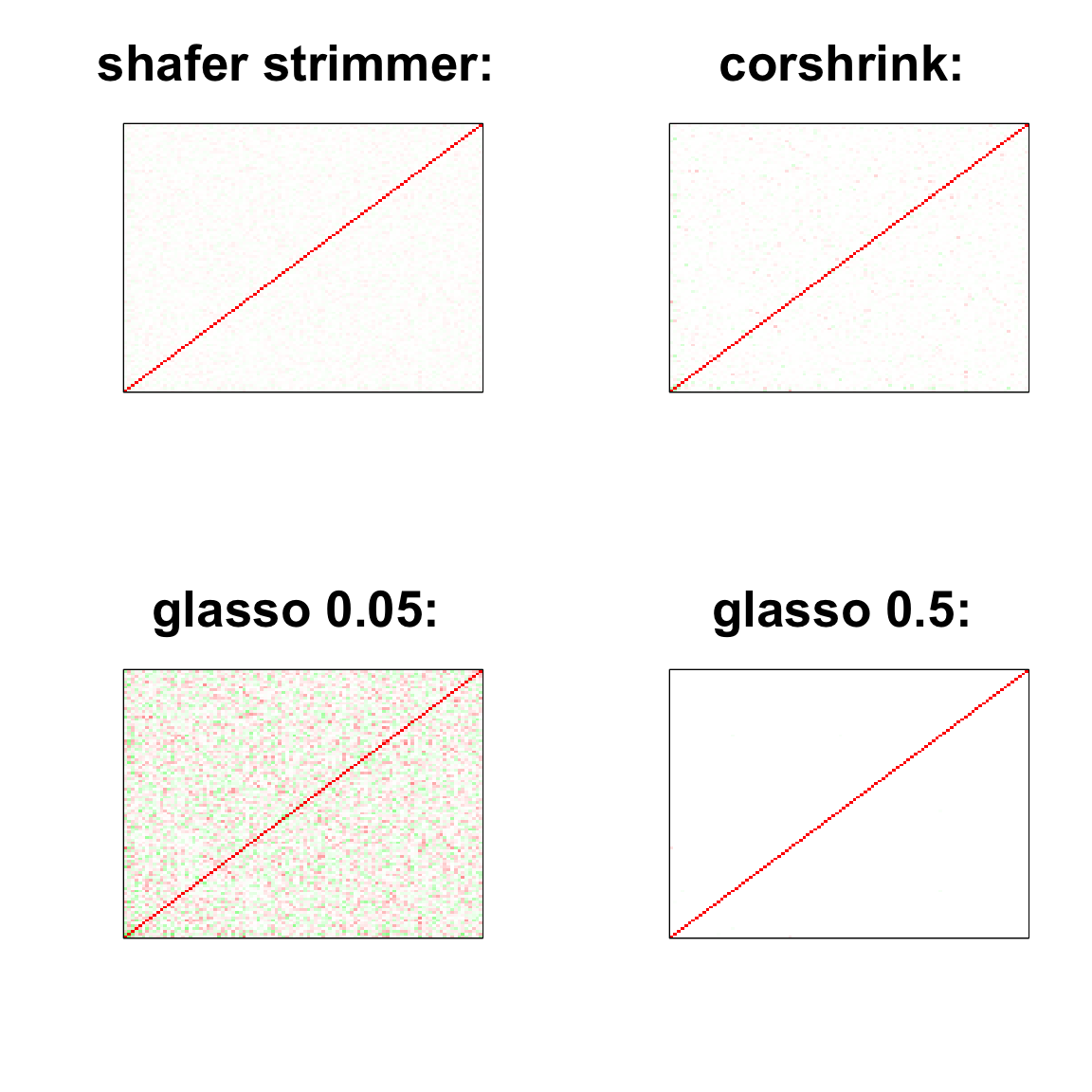
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(spcor_mat),
col=col, main=paste0("spcov:", expression(lambda), "=", lam), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
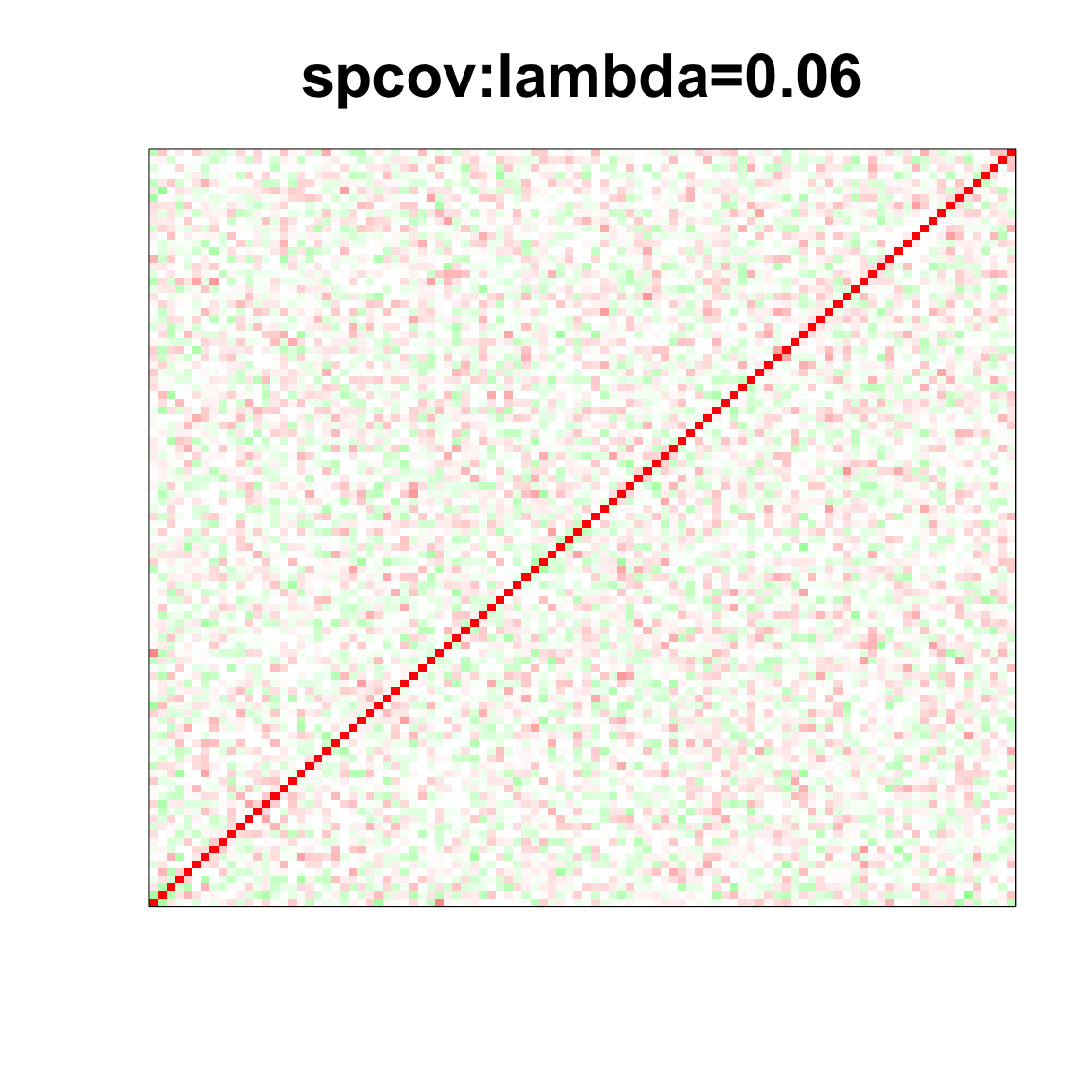
n = 5, P=100
n <- 5
P <- 100
ll <- DM_graph(n,P,g_type = "scale-free")
## Generating data from the multivariate normal distribution with the scale-free graph structure....
## done.
data <- rbind(ll$Xtrain, ll$Xtest)
Sigma <- ll$Sigma
corSigma <- cov2cor(Sigma)
Pat <- matrix(1, P, P)
diag(Pat) <- 0
lam <- 0.06
step.size <- 100
tol <- 1e-4
covmat <- cov(data)
mm <- spcov(Sigma=covmat+0.1*diag(1,P), S=covmat+0.1*diag(1,P), lambda=lam * Pat,step.size=step.size, n.inner.steps=200, thr.inner=0, tol.outer=tol, trace=1)
## ---
## using Nesterov, backtracking line search
## ---
## objective: 132.092
## step size given to GGDescent/Nesterov: 100
## objective: -56.55613 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 20
## step size given to GGDescent/Nesterov: 20
## objective: -60.26477 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 4
## step size given to GGDescent/Nesterov: 4
## objective: -60.28042 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.8
## step size given to GGDescent/Nesterov: 0.8
## objective: -60.28227 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.16
## step size given to GGDescent/Nesterov: 0.16
## objective: -60.28274 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.032
## step size given to GGDescent/Nesterov: 0.032
## objective: -60.28286 ( 200 iterations, max step size: 0.00128 )
## step size given to GGDescent/Nesterov: 0.032
## objective: -60.2829 ( 200 iterations, max step size: 0.00128 )
## MM converged in 7 steps!
spcor_mat <- cov2cor(mm$Sigma)
#devtools::install_github("kkdey/CorShrink")
#library(CorShrink)
#sessionInfo()
cov_mat <- cov(data);
system.time(strimmer_sample <- corpcor::cov.shrink(data))
## Estimating optimal shrinkage intensity lambda.var (variance vector): 0.8478
##
## Estimating optimal shrinkage intensity lambda (correlation matrix): 0.7522
## user system elapsed
## 0.002 0.000 0.002
system.time(glasso_sample_005 <- glasso::glasso(cov_mat, rho = 0.05))
## user system elapsed
## 0.663 0.002 0.667
system.time(glasso_sample_05 <- glasso::glasso(cov_mat, rho = 0.5))
## user system elapsed
## 0.059 0.000 0.059
system.time(glasso_sample_1 <- glasso::glasso(cov_mat, rho = 1))
## user system elapsed
## 0.012 0.000 0.013
system.time(glasso_sample_10 <- glasso::glasso(cov_mat, rho = 10))
## user system elapsed
## 0.002 0.000 0.001
system.time(cov_sample_ML <- CorShrinkMatrix(cov2cor(cov_mat), nsamp = matrix(n, P, P), ash.control = list(mixcompdist = "normal", nullweight = 10)))
## ash cor only and ash cor PD matrices are different
## user system elapsed
## 1.758 0.033 1.798
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(cov_mat)),
col=col, main=paste0("sample corr: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
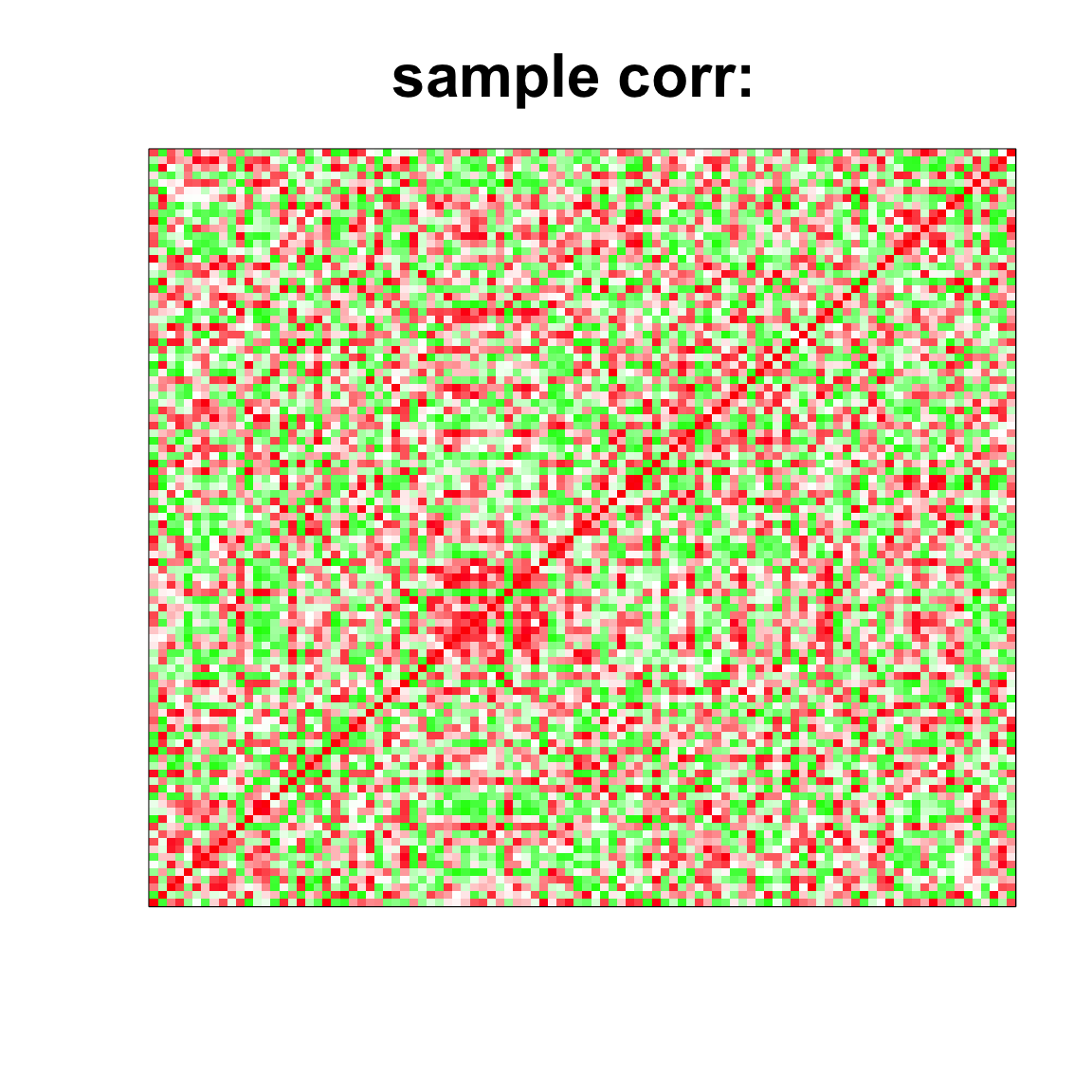
par(mfrow = c(2,2))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(strimmer_sample)),
col=col, main=paste0("shafer strimmer: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov_sample_ML$ash_cor_only),
col=col, main=paste0("corshrink: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(glasso_sample_005$w)),
col=col, main=paste0("glasso 0.05: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(glasso_sample_05$w)),
col=col, main=paste0("glasso 0.5: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
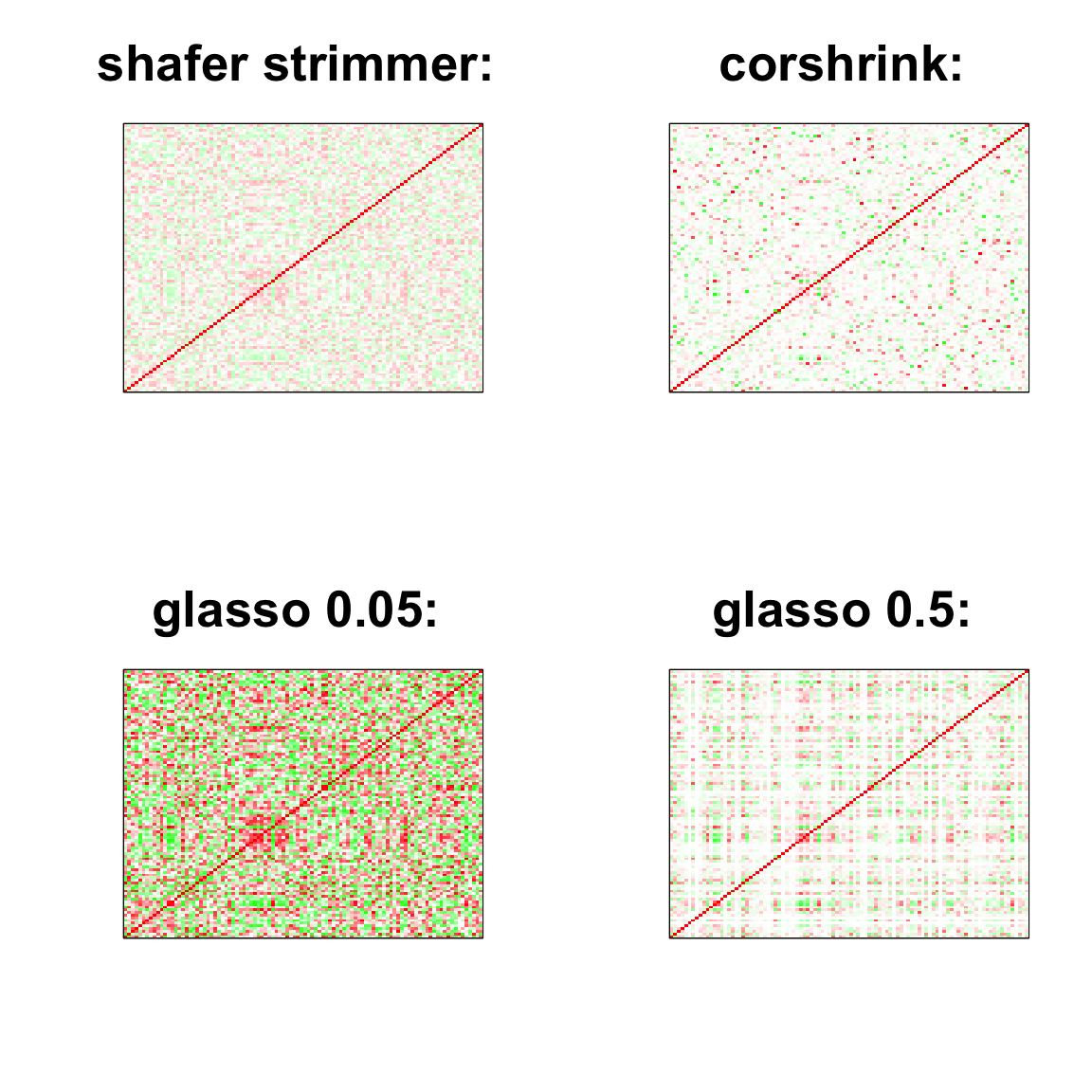
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(spcor_mat),
col=col, main=paste0("spcov:", expression(lambda), "=", lam), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))