CorShrink on dsc simulated data (DM diagonal)
Kushal K Dey
6/21/2017
library(spcov)
library(CorShrink)
library(corpcor)
library(glasso)DM_diagonal = function(n,P){
library("MASS")
index1=sort(sample(seq(1:n),(n/2)))
index2=seq(1:n)[-index1]
Sigma = diag(rchisq(P,3))
data = mvrnorm(n,rep(0,P),Sigma)
Xtest = data[index2,]
Xtrain = data[index1,]
Omega = diag((1/rchisq(P,3)))
return(list(Xtrain = Xtrain, Xtest = Xtest, Omega = Omega))
}n = 10, P=100
n <- 10
P <- 100
ll <- DM_diagonal(n=n, P=P)
data <- rbind(ll$Xtrain, ll$Xtest)
Sigma <- solve(ll$Omega)
corSigma <- cov2cor(Sigma)Pat <- matrix(1, P, P)
diag(Pat) <- 0lam <- 0.06
step.size <- 100
tol <- 1e-4
covmat <- cov(data)
mm <- spcov(Sigma=covmat+0.1*diag(1,P), S=covmat+0.1*diag(1,P), lambda=lam * Pat,step.size=step.size, n.inner.steps=200, thr.inner=0, tol.outer=tol, trace=1)## ---
## using Nesterov, backtracking line search
## ---
## objective: 316.7115
## step size given to GGDescent/Nesterov: 100
## objective: 17.8547 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 20
## step size given to GGDescent/Nesterov: 20
## objective: 8.003283 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 4
## step size given to GGDescent/Nesterov: 4
## objective: 7.939534 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.8
## step size given to GGDescent/Nesterov: 0.8
## objective: 7.935671 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.16
## step size given to GGDescent/Nesterov: 0.16
## objective: 7.934714 ( 200 iterations, max step size: 0.00128 )
## Reducing step size to 0.032
## step size given to GGDescent/Nesterov: 0.032
## objective: 7.934457 ( 200 iterations, max step size: 0.00128 )
## step size given to GGDescent/Nesterov: 0.032
## objective: 7.934387 ( 200 iterations, max step size: 0.00128 )
## MM converged in 7 steps!spcor_mat <- cov2cor(mm$Sigma)#devtools::install_github("kkdey/CorShrink")
#library(CorShrink)
#sessionInfo()
cov_mat <- cov(data);
system.time(strimmer_sample <- corpcor::cov.shrink(data))## Estimating optimal shrinkage intensity lambda.var (variance vector): 0.2749
##
## Estimating optimal shrinkage intensity lambda (correlation matrix): 0.8658## user system elapsed
## 0.020 0.001 0.021system.time(glasso_sample_005 <- glasso::glasso(cov_mat, rho = 0.05))## user system elapsed
## 0.844 0.002 0.851system.time(glasso_sample_05 <- glasso::glasso(cov_mat, rho = 0.5))## user system elapsed
## 0.133 0.000 0.134system.time(glasso_sample_1 <- glasso::glasso(cov_mat, rho = 1))## user system elapsed
## 0.048 0.000 0.049system.time(glasso_sample_10 <- glasso::glasso(cov_mat, rho = 10))## user system elapsed
## 0.001 0.000 0.002system.time(cov_sample_ML <- CorShrinkMatrix(cov2cor(cov_mat), nsamp = matrix(n, P, P),
ash.control = list(mixcompdist = "normal", nullweight = 100)))## ash cor only and ash cor PD matrices are different## user system elapsed
## 1.445 0.043 1.496col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(cov_mat)),
col=col, main=paste0("sample corr: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))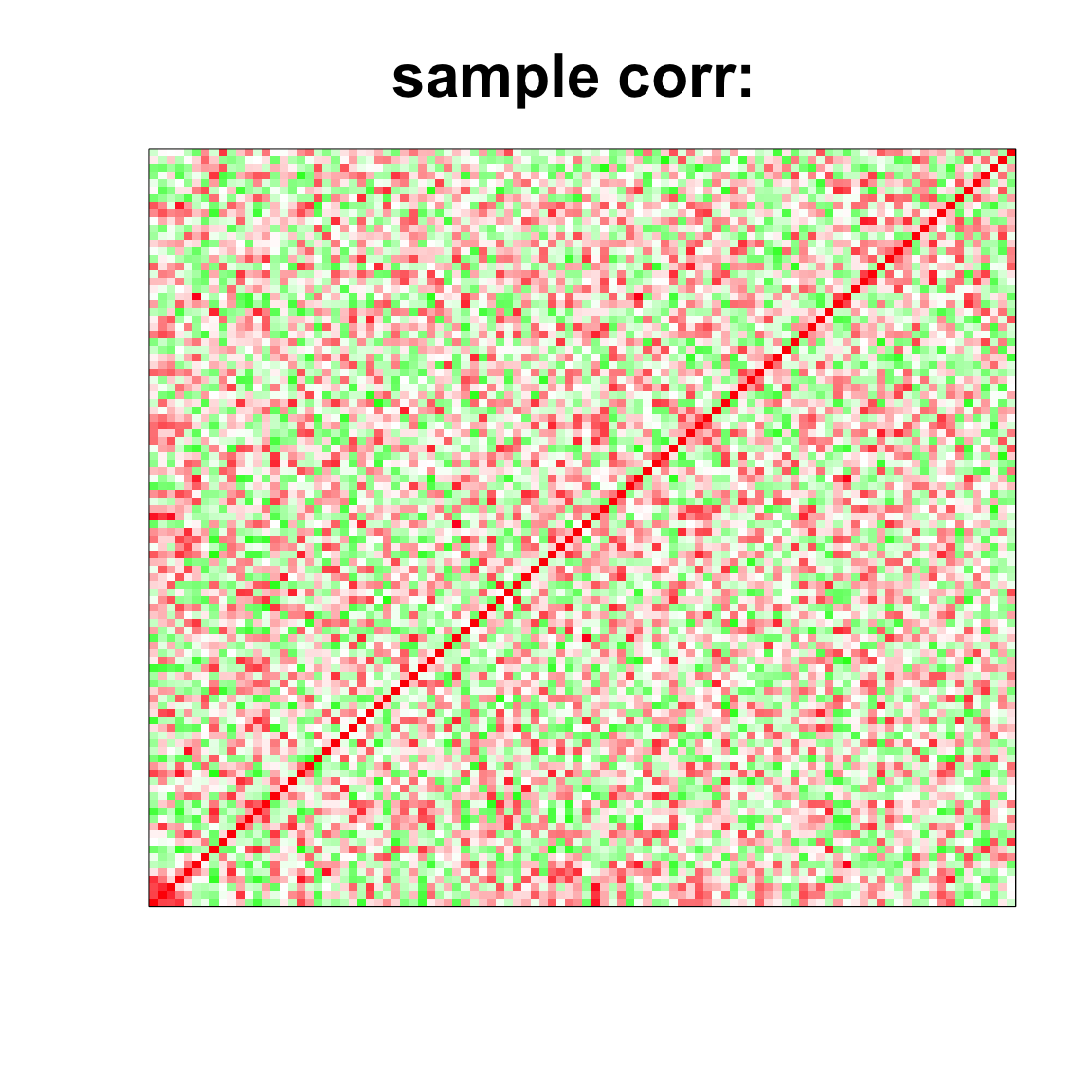
par(mfrow=c(2,2))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(strimmer_sample)),
col=col, main=paste0("shafer strimmer: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov_sample_ML$ash_cor_PD),
col=col, main=paste0("corshrink: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(glasso_sample_005$w)),
col=col, main=paste0("glasso 0.05: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(glasso_sample_05$w)),
col=col, main=paste0("glasso 0.5: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))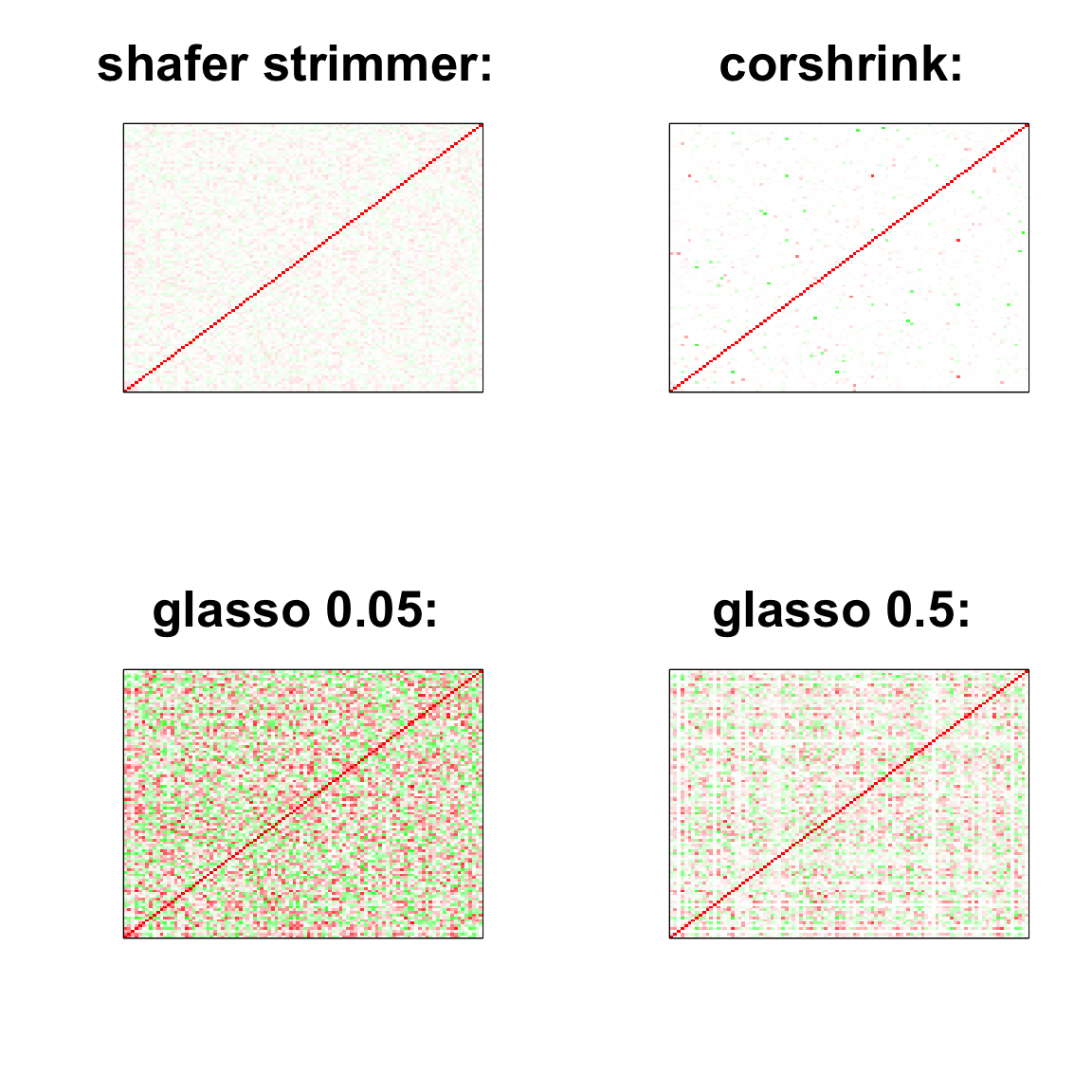
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(spcor_mat),
col=col, main=paste0("spcov:", expression(lambda), "=", lam), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))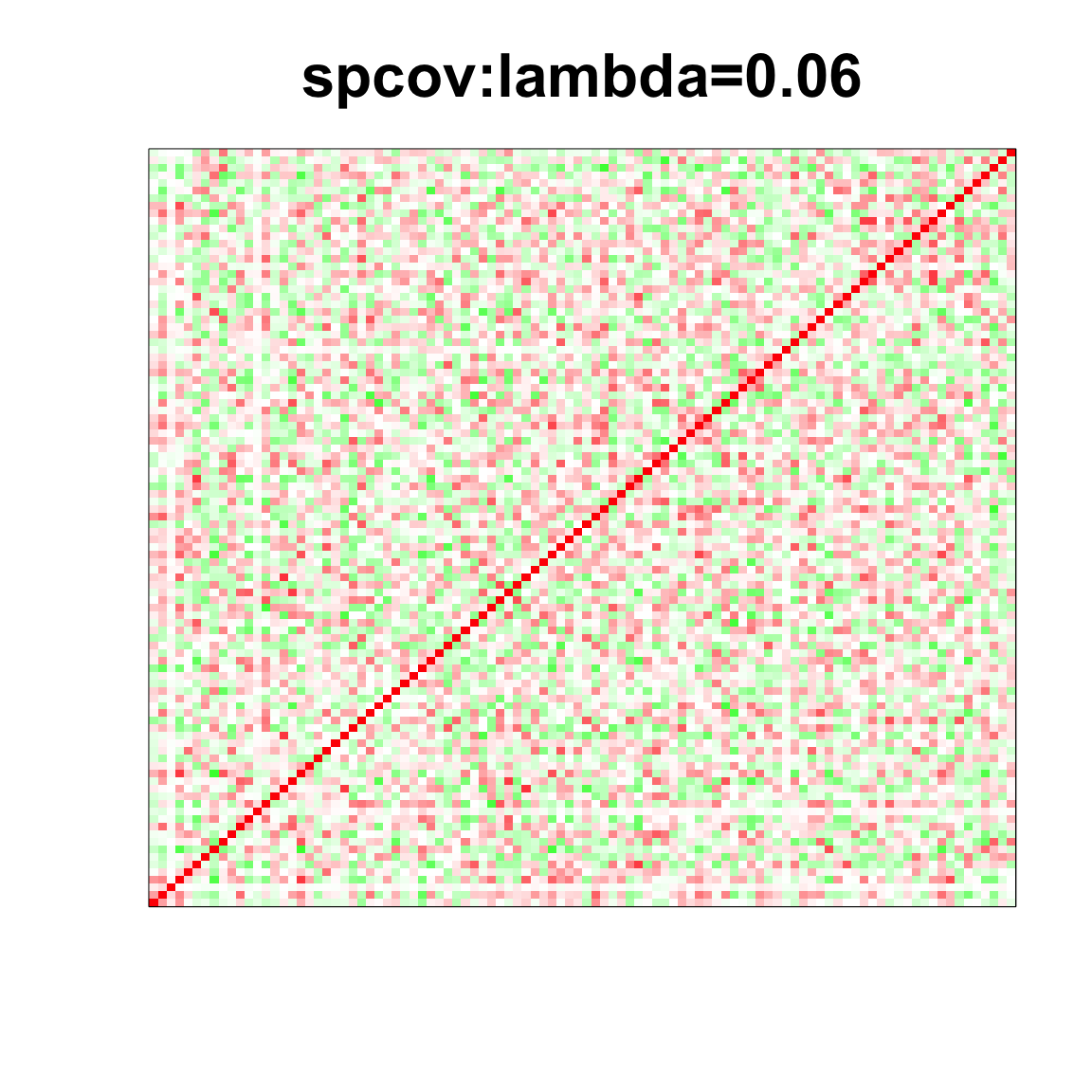
n = 50, P=100
n <- 50
P <- 100
ll <- DM_diagonal(n=n, P=P)
data <- rbind(ll$Xtrain, ll$Xtest)
Sigma <- solve(ll$Omega)
corSigma <- cov2cor(Sigma)Pat <- matrix(1, P, P)
diag(Pat) <- 0lam <- 0.06
step.size <- 100
tol <- 1e-4
covmat <- cov(data)
mm <- spcov(Sigma=covmat+0.1*diag(1,P), S=covmat+0.1*diag(1,P), lambda=lam * Pat,step.size=step.size, n.inner.steps=200, thr.inner=0, tol.outer=tol, trace=1)## ---
## using Nesterov, backtracking line search
## ---
## objective: 214.1995
## step size given to GGDescent/Nesterov: 100
## objective: 125.3437 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 20
## step size given to GGDescent/Nesterov: 20
## objective: 122.9463 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 4
## step size given to GGDescent/Nesterov: 4
## objective: 122.8393 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 0.8
## step size given to GGDescent/Nesterov: 0.8
## objective: 122.8232 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 0.16
## step size given to GGDescent/Nesterov: 0.16
## objective: 122.8184 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 0.032
## step size given to GGDescent/Nesterov: 0.032
## objective: 122.8166 ( 200 iterations, max step size: 0.0064 )
## step size given to GGDescent/Nesterov: 0.032
## objective: 122.8158 ( 200 iterations, max step size: 0.0064 )
## step size given to GGDescent/Nesterov: 0.032
## objective: 122.8155 ( 200 iterations, max step size: 0.0064 )
## step size given to GGDescent/Nesterov: 0.032
## objective: 122.8153 ( 200 iterations, max step size: 0.0064 )
## step size given to GGDescent/Nesterov: 0.032
## objective: 122.8152 ( 200 iterations, max step size: 0.0064 )
## MM converged in 10 steps!spcor_mat <- cov2cor(mm$Sigma)#devtools::install_github("kkdey/CorShrink")
#library(CorShrink)
#sessionInfo()
cov_mat <- cov(data);
system.time(strimmer_sample <- corpcor::cov.shrink(data))## Estimating optimal shrinkage intensity lambda.var (variance vector): 0.0773
##
## Estimating optimal shrinkage intensity lambda (correlation matrix): 1## user system elapsed
## 0.079 0.001 0.082system.time(glasso_sample_005 <- glasso::glasso(cov_mat, rho = 0.05))## user system elapsed
## 0.352 0.001 0.354system.time(glasso_sample_05 <- glasso::glasso(cov_mat, rho = 0.5))## user system elapsed
## 0.028 0.000 0.028system.time(glasso_sample_1 <- glasso::glasso(cov_mat, rho = 1))## user system elapsed
## 0.008 0.000 0.008system.time(glasso_sample_10 <- glasso::glasso(cov_mat, rho = 10))## user system elapsed
## 0.001 0.000 0.001system.time(cov_sample_ML <- CorShrinkMatrix(cov2cor(cov_mat), nsamp = matrix(n, P, P), ash.control = list(mixcompdist = "normal", nullweight = 100)))## ash cor only and ash cor PD matrices are different## user system elapsed
## 1.885 0.027 1.924col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(cov_mat)),
col=col, main=paste0("sample corr: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))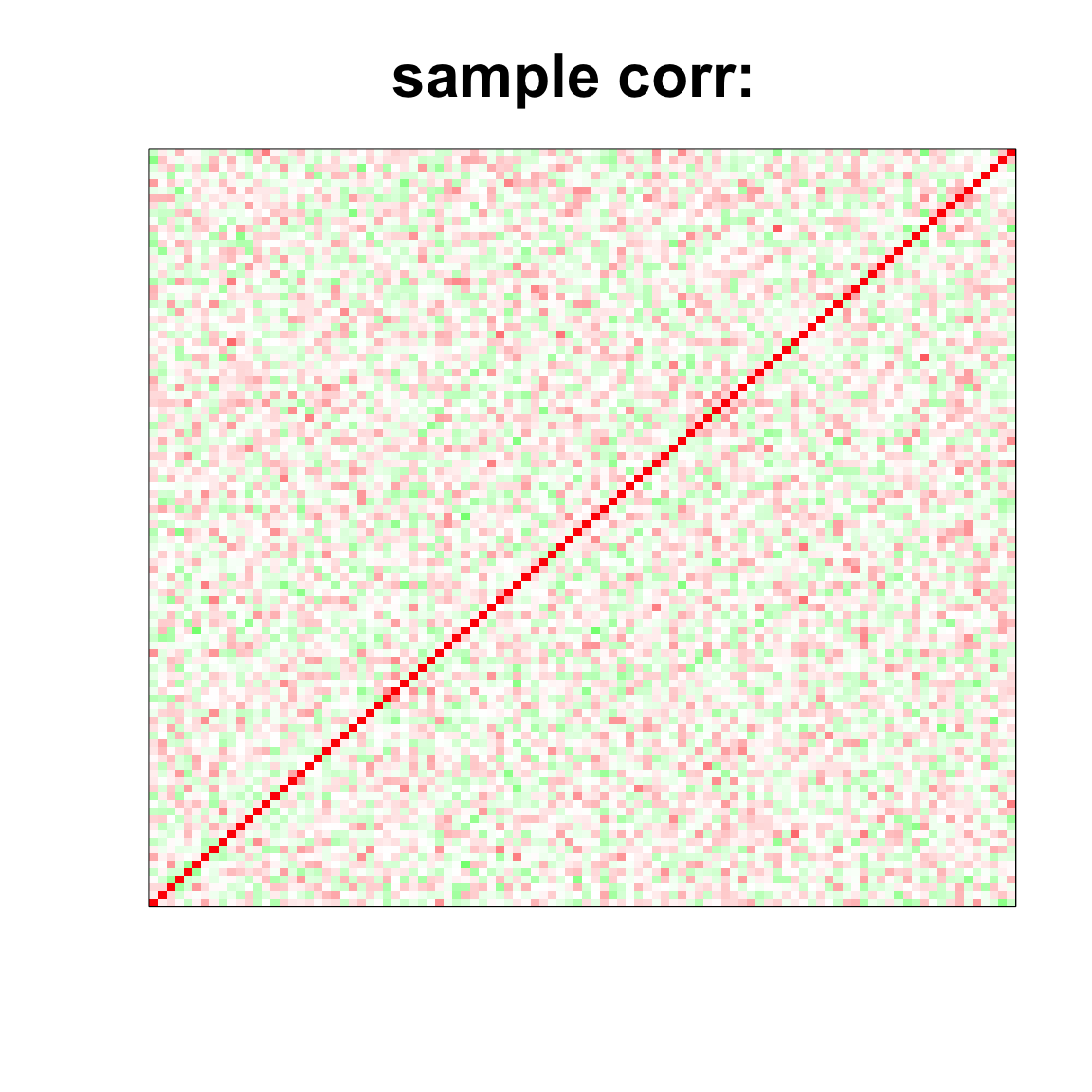
par(mfrow = c(2,2))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(strimmer_sample)),
col=col, main=paste0("shafer strimmer: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov_sample_ML$ash_cor_only),
col=col, main=paste0("corshrink: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(glasso_sample_005$w)),
col=col, main=paste0("glasso 0.05: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(glasso_sample_05$w)),
col=col, main=paste0("glasso 0.5: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))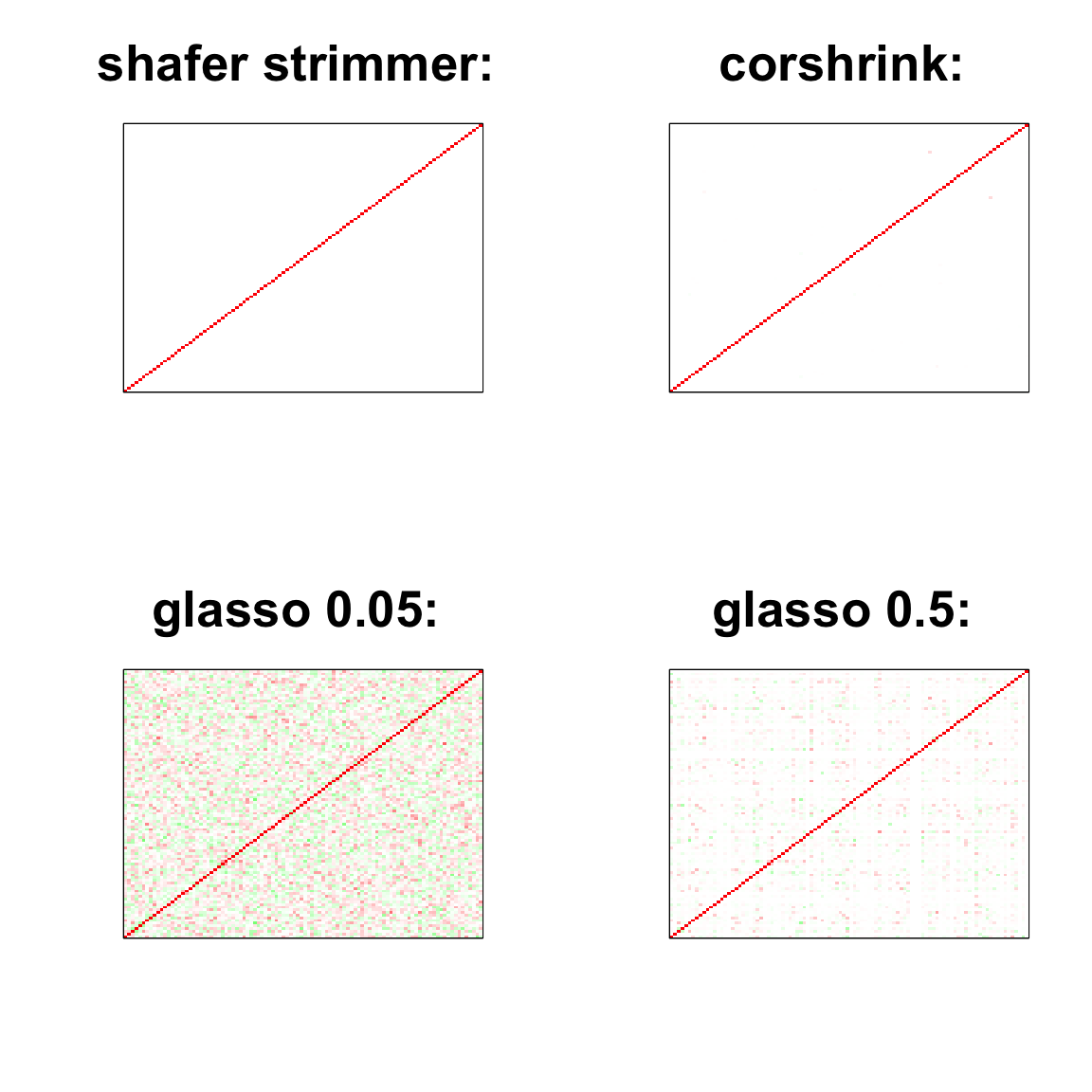
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(spcor_mat),
col=col, main=paste0("spcov:", expression(lambda), "=", lam), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))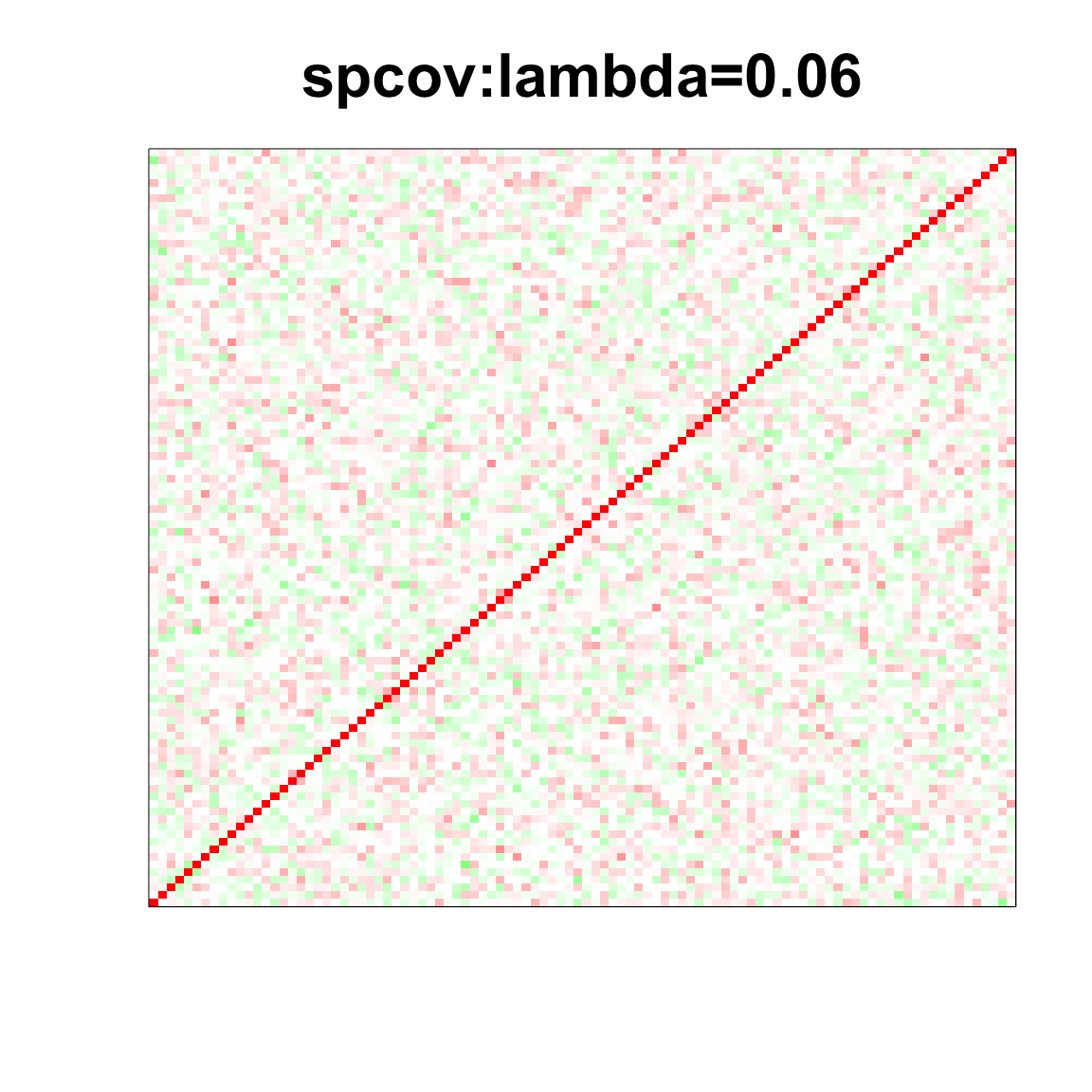
n = 5, P=100
n <- 5
P <- 100
ll <- DM_diagonal(n=n, P=P)
data <- rbind(ll$Xtrain, ll$Xtest)
Sigma <- solve(ll$Omega)
corSigma <- cov2cor(Sigma)Pat <- matrix(1, P, P)
diag(Pat) <- 0lam <- 0.06
step.size <- 100
tol <- 1e-4
covmat <- cov(data)
mm <- spcov(Sigma=covmat+0.1*diag(1,P), S=covmat+0.1*diag(1,P), lambda=lam * Pat,step.size=step.size, n.inner.steps=200, thr.inner=0, tol.outer=tol, trace=1)## ---
## using Nesterov, backtracking line search
## ---
## objective: 423.3422
## step size given to GGDescent/Nesterov: 100
## objective: 21.93159 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 20
## step size given to GGDescent/Nesterov: 20
## objective: -25.02688 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 4
## step size given to GGDescent/Nesterov: 4
## objective: -27.62124 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 0.8
## step size given to GGDescent/Nesterov: 0.8
## objective: -27.66326 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 0.16
## step size given to GGDescent/Nesterov: 0.16
## objective: -27.6744 ( 200 iterations, max step size: 0.0064 )
## Reducing step size to 0.032
## step size given to GGDescent/Nesterov: 0.032
## objective: -27.67749 ( 200 iterations, max step size: 0.00128 )
## step size given to GGDescent/Nesterov: 0.032
## objective: -27.67833 ( 200 iterations, max step size: 0.00128 )
## step size given to GGDescent/Nesterov: 0.032
## objective: -27.67856 ( 200 iterations, max step size: 0.00128 )
## step size given to GGDescent/Nesterov: 0.032
## objective: -27.67863 ( 200 iterations, max step size: 0.00128 )
## MM converged in 9 steps!spcor_mat <- cov2cor(mm$Sigma)#devtools::install_github("kkdey/CorShrink")
#library(CorShrink)
#sessionInfo()
cov_mat <- cov(data);
system.time(strimmer_sample <- corpcor::cov.shrink(data))## Estimating optimal shrinkage intensity lambda.var (variance vector): 0.4583
##
## Estimating optimal shrinkage intensity lambda (correlation matrix): 0.7542## user system elapsed
## 0.009 0.000 0.009system.time(glasso_sample_005 <- glasso::glasso(cov_mat, rho = 0.05))## user system elapsed
## 0.965 0.003 0.972system.time(glasso_sample_05 <- glasso::glasso(cov_mat, rho = 0.5))## user system elapsed
## 0.144 0.000 0.145system.time(glasso_sample_1 <- glasso::glasso(cov_mat, rho = 1))## user system elapsed
## 0.068 0.000 0.068system.time(glasso_sample_10 <- glasso::glasso(cov_mat, rho = 10))## user system elapsed
## 0.002 0.000 0.001system.time(cov_sample_ML <- CorShrinkMatrix(cov2cor(cov_mat), nsamp = matrix(n, P, P), ash.control = list(mixcompdist = "normal", nullweight = 100)))## ash cor only and ash cor PD matrices are different## user system elapsed
## 1.202 0.041 1.250col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(cov_mat)),
col=col, main=paste0("sample corr: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))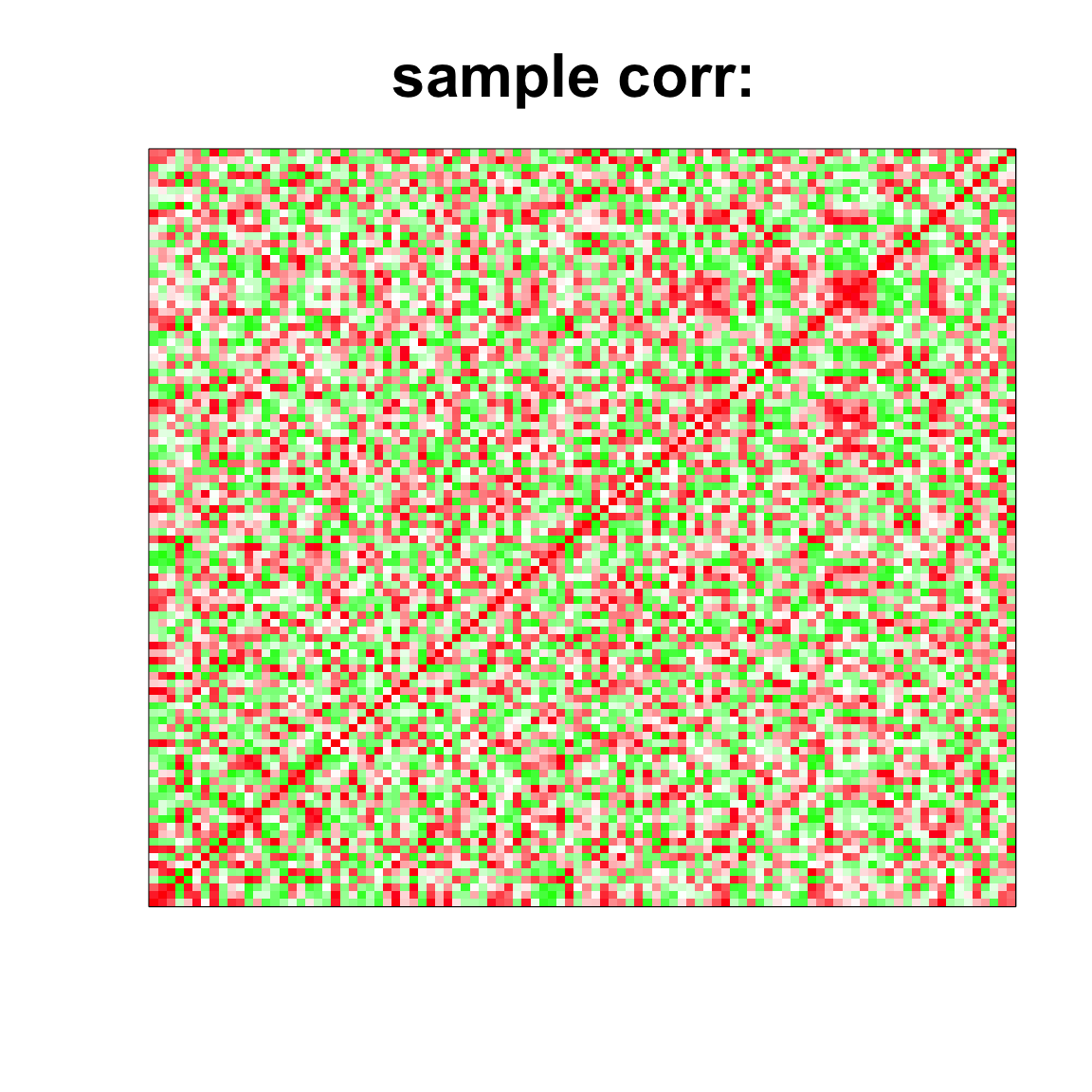
par(mfrow = c(2,2))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(strimmer_sample)),
col=col, main=paste0("shafer strimmer: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov_sample_ML$ash_cor_only),
col=col, main=paste0("corshrink: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(glasso_sample_005$w)),
col=col, main=paste0("glasso 0.05: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(cov2cor(glasso_sample_05$w)),
col=col, main=paste0("glasso 0.5: "), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))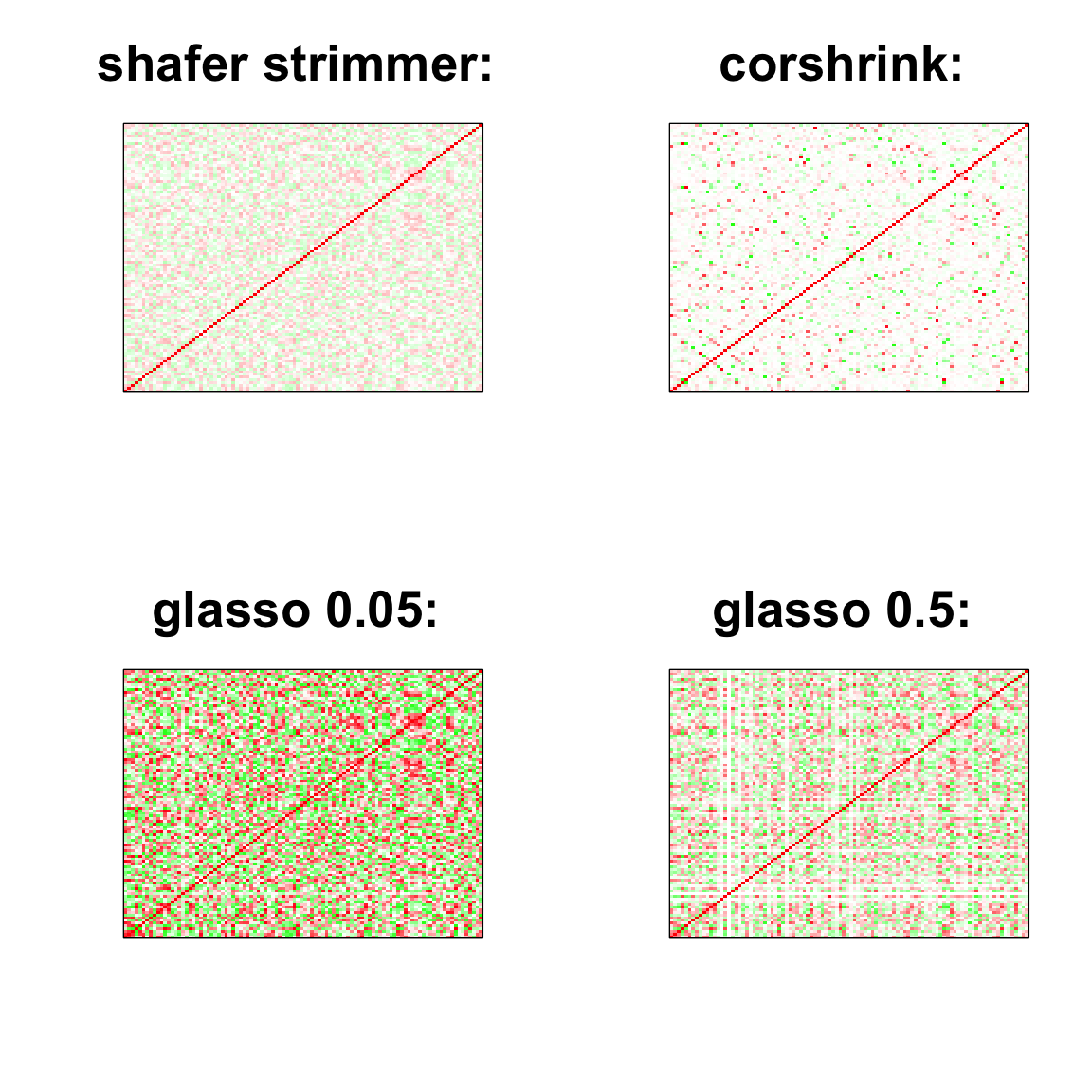
col=c(rev(rgb(seq(1,0,length=1000),1,seq(1,0,length=1000))),
rgb(1,seq(1,0,length=1000),seq(1,0,length=1000)))
image(as.matrix(spcor_mat),
col=col, main=paste0("spcov:", expression(lambda), "=", lam), cex.main=2,
xaxt = "n", yaxt = "n", zlim=c(-1,1))
This R Markdown site was created with workflowr