Wallacea region GoM analysis - with seabirds
Kushal K Dey
3/25/2018
Intro
Here we observe the presence absence data of bird species in the Australasian region (Wallacea). We try to interpret that in the context of our Grade of Membership (GoM) model and its applications to presence absence data.
Packages
library(methClust)
library(CountClust)
library(rasterVis)
library(gtools)
library(sp)
library(rgdal)
library(ggplot2)
library(maps)
library(mapdata)
library(mapplots)
library(scales)
library(ggthemes)Load the data
Wallacea Region data
birds <- get(load("../data/wallacea_birds.rda"))
latlong_chars_birds <- rownames(birds)
latlong <- cbind.data.frame(as.numeric(sapply(latlong_chars_birds,
function(x) return(strsplit(x, "_")[[1]][1]))),
as.numeric(sapply(latlong_chars_birds,
function(x) return(strsplit(x, "_")[[1]][2]))))Map of Wallacea
world_map <- map_data("world")
world_map <- world_map[world_map$region != "Antarctica",] # intercourse antarctica
world_map <- world_map[world_map$long > 90 & world_map$long < 160, ]
world_map <- world_map[world_map$lat > -18 & world_map$lat < 20, ]
p <- ggplot() + coord_fixed() +
xlab("") + ylab("")
#Add map to base plot
base_world_messy <- p + geom_polygon(data=world_map, aes(x=long, y=lat, group=group), colour="light green", fill="light green")
cleanup <-
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
panel.background = element_rect(fill = 'white', colour = 'white'),
axis.line = element_line(colour = "white"), legend.position="none",
axis.ticks=element_blank(), axis.text.x=element_blank(),
axis.text.y=element_blank())
base_world <- base_world_messy + cleanup
base_world
richness <- rowSums(birds)colorGradient <- colorRampPalette(c("blue", "white", "red"))(max(richness))
plot(latlong[,1], latlong[,2], col= colorGradient[richness], pch = 20, cex = 1.5)
GoM model
Applying methclust presence absence Grade of Membership model to the presence absence data
topics_clust <- list()
topics_clust[[1]] <- NULL
for(k in 2:10){
topics_clust[[k]] <- meth_topics(birds, 1 - birds,
K=k, tol = 10, use_squarem = FALSE)
}
save(topics_clust, file = "../output/methClust_wallacea_birds.rda")Visualization
topics_clust <- get(load("../output/methClust_wallacea_birds.rda"))color = c("red", "cornflowerblue", "cyan", "brown4", "burlywood", "darkgoldenrod1",
"azure4", "green","deepskyblue","yellow", "azure1")
intensity <- 0.8
for(k in 2:10){
png(filename=paste0("../docs/Wallacea_w_seabirds/geostructure_birds_", k, ".png"),width = 1000, height = 800)
map("worldHires",
ylim=c(-18,20), xlim=c(90,160), # Re-defines the latitude and longitude range
col = "gray", fill=TRUE, mar=c(0.1,0.1,0.1,0.1))
lapply(1:dim(topics_clust[[k]]$omega)[1], function(r)
add.pie(z=as.integer(100*topics_clust[[k]]$omega[r,]),
x=latlong[r,1], y=latlong[r,2], labels=c("","",""),
radius = 0.5,
col=c(alpha(color[1],intensity),alpha(color[2],intensity),
alpha(color[3], intensity), alpha(color[4], intensity),
alpha(color[5], intensity), alpha(color[6], intensity),
alpha(color[7], intensity), alpha(color[8], intensity),
alpha(color[9], intensity), alpha(color[10], intensity),
alpha(color[11], intensity))));
dev.off()
}The geostructure plot for different K.
K = 2
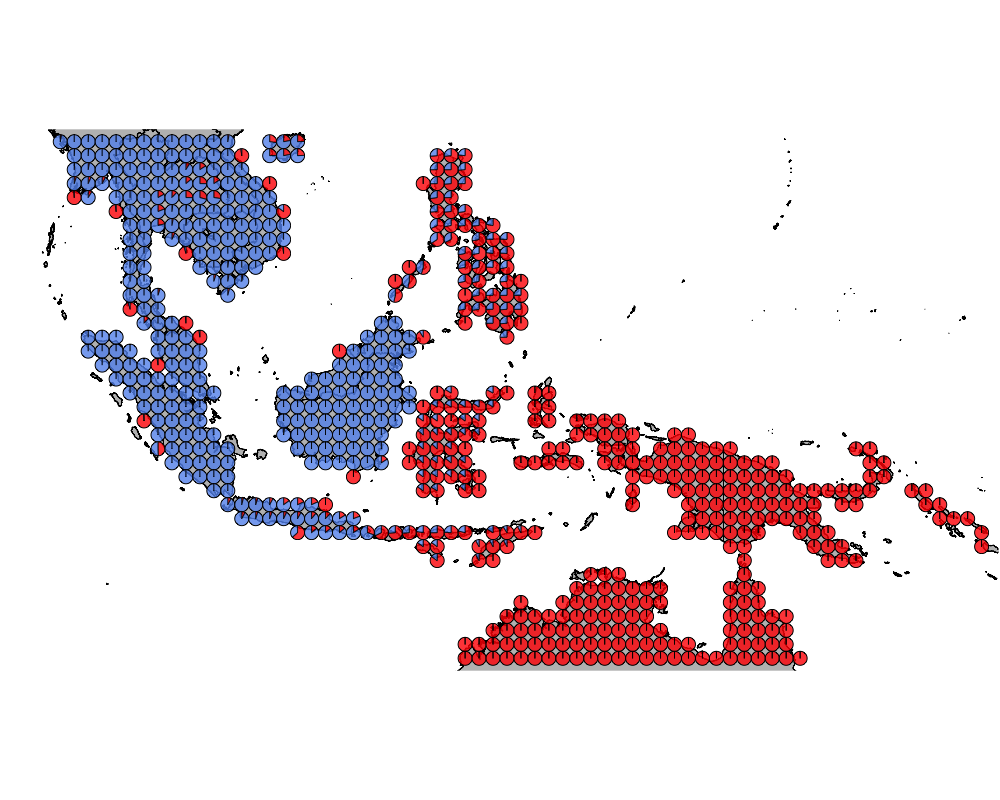
geostructure2
K = 3
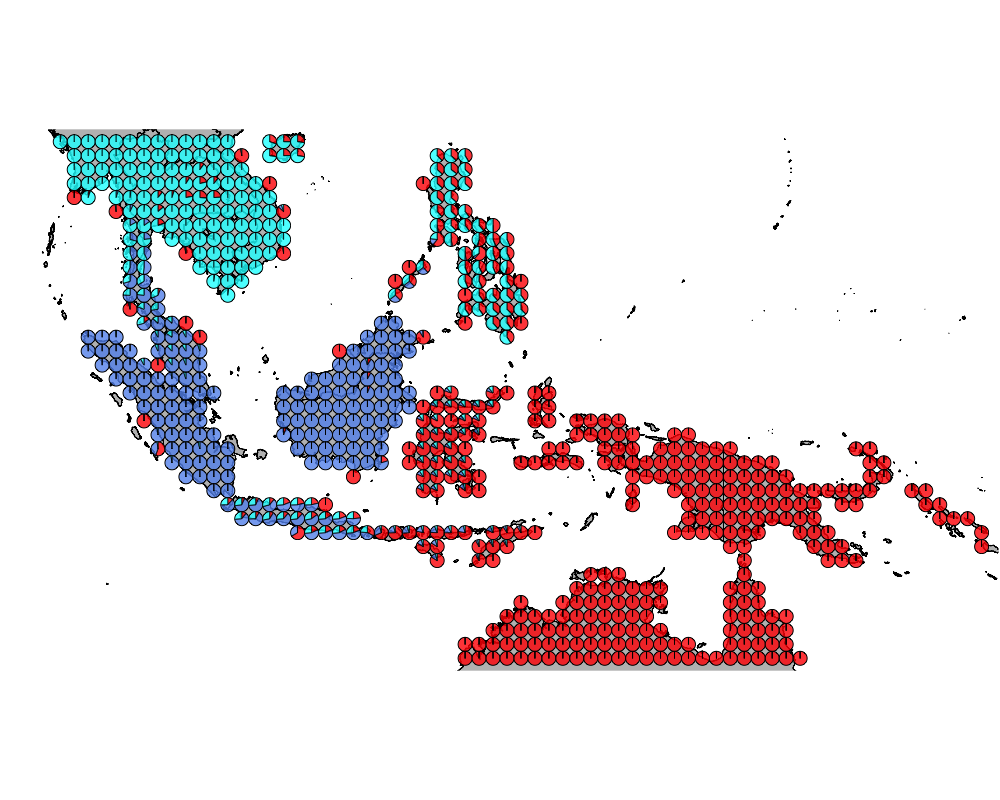
geostructure3
K = 4
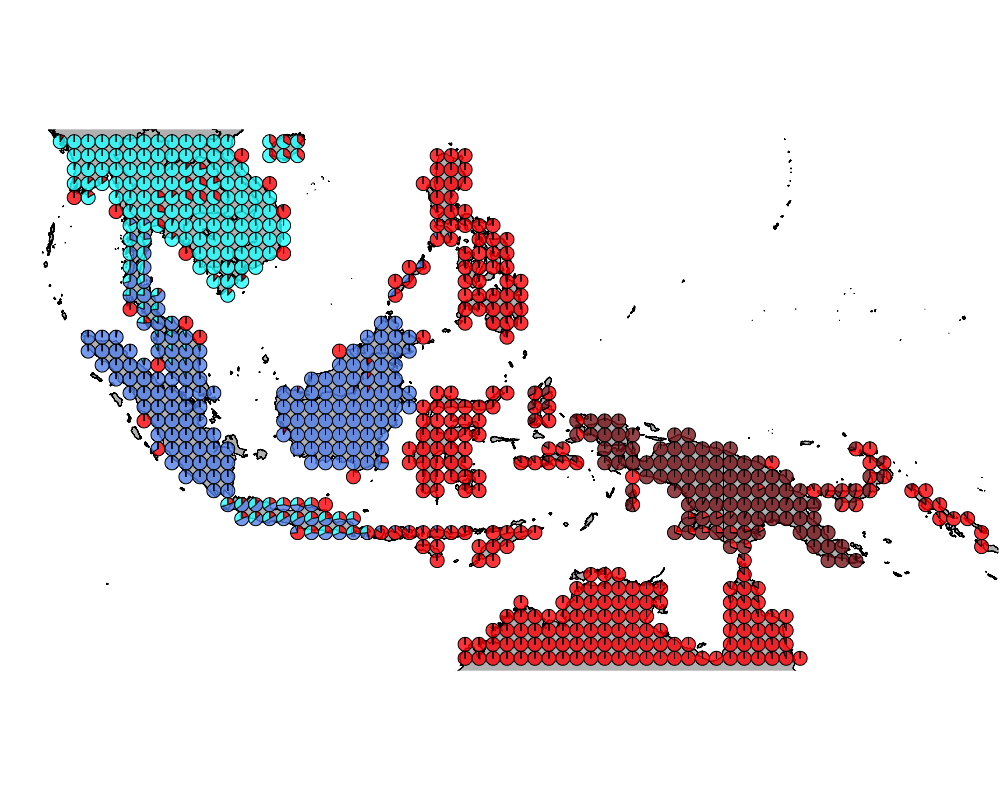
geostructure4
K = 5
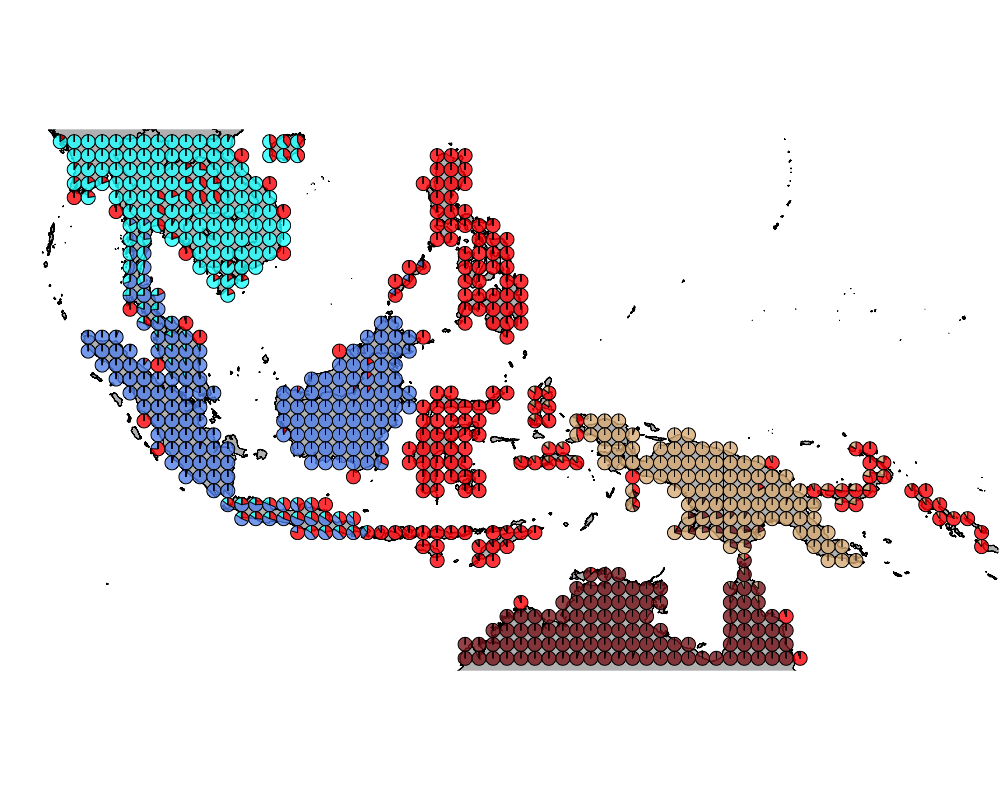
geostructure5
K = 6
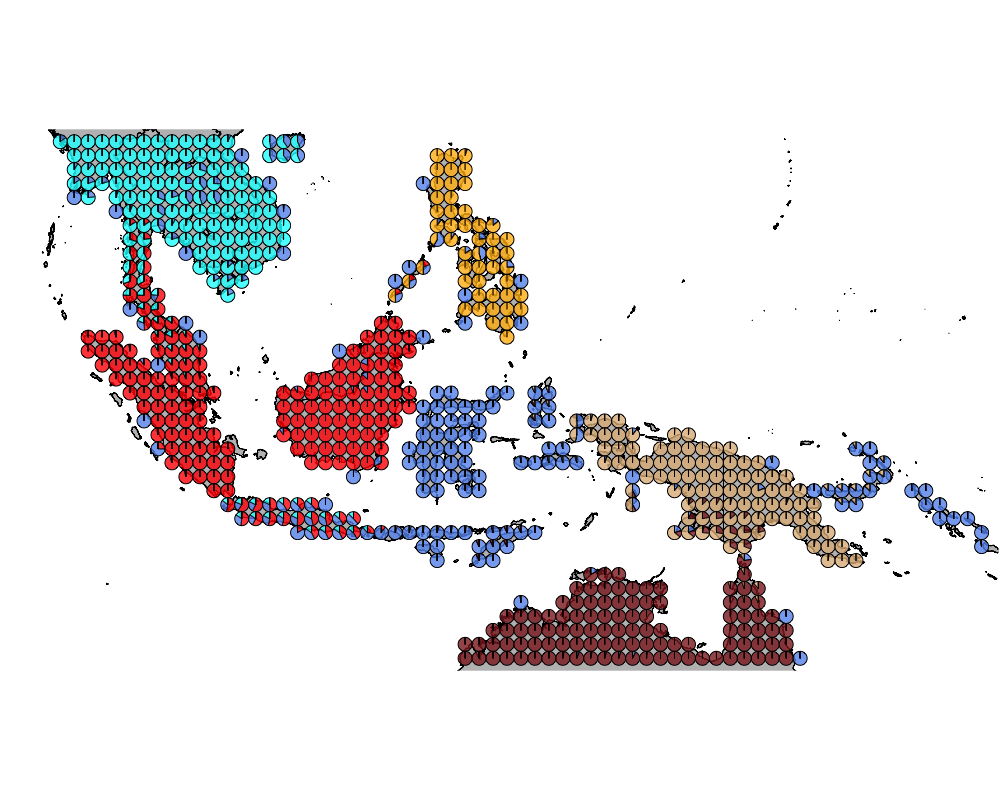
geostructure6
K = 7
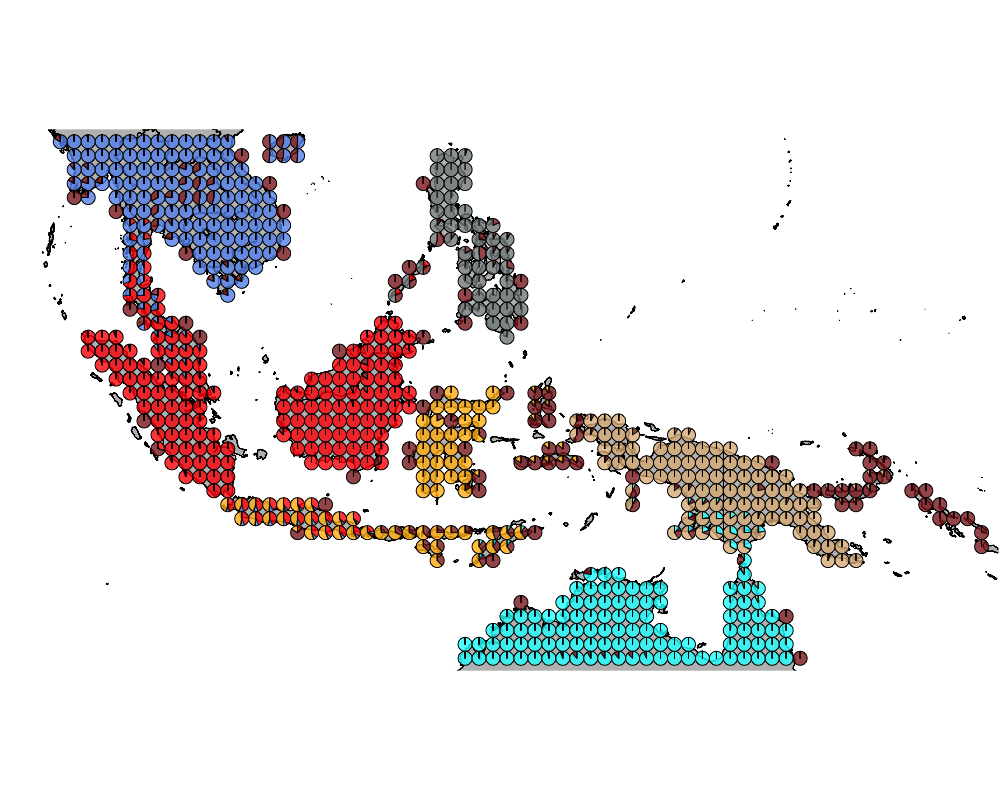
geostructure7
K = 8
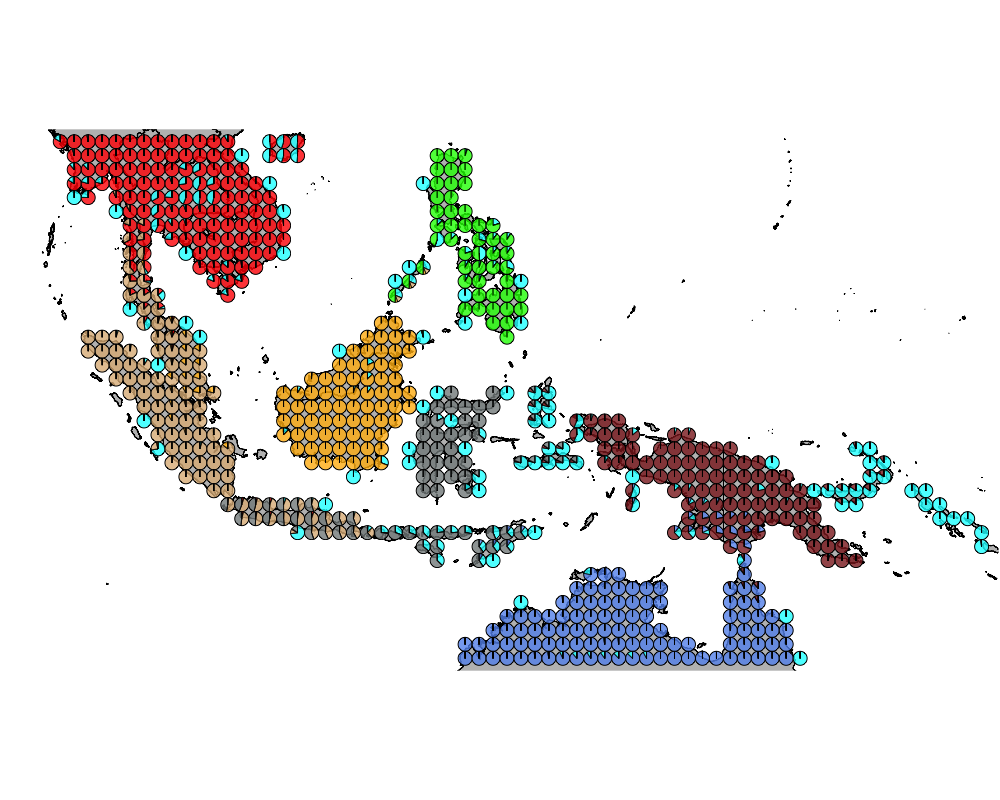
geostructure8
K = 9
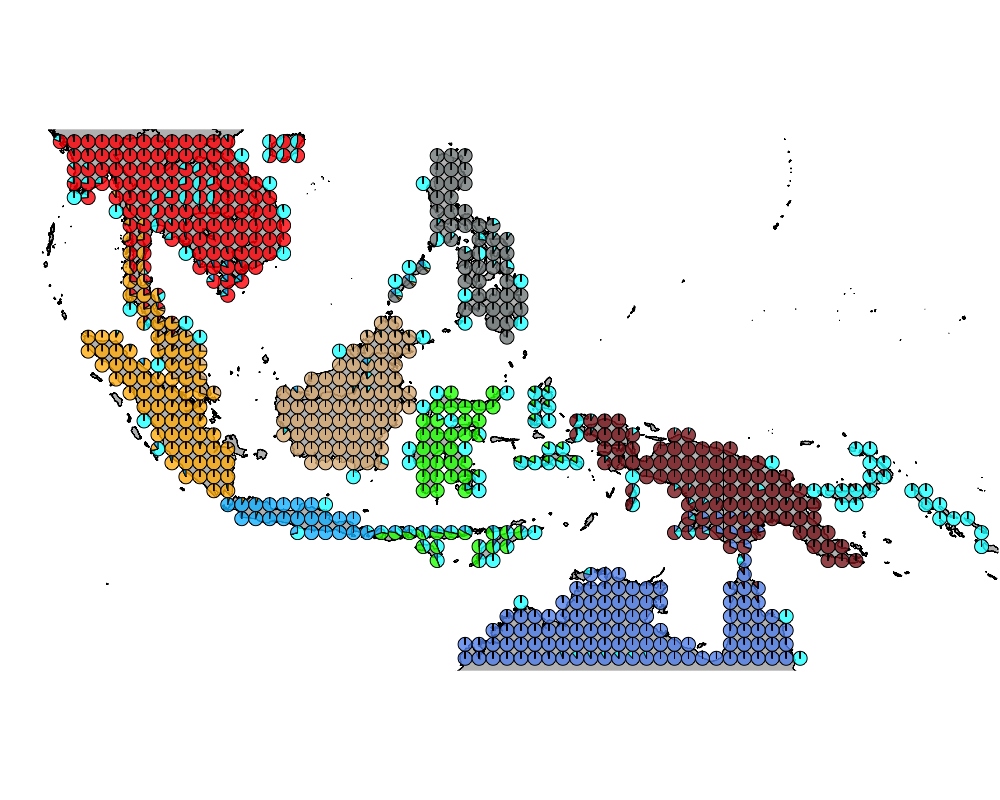
geostructure9
K = 10
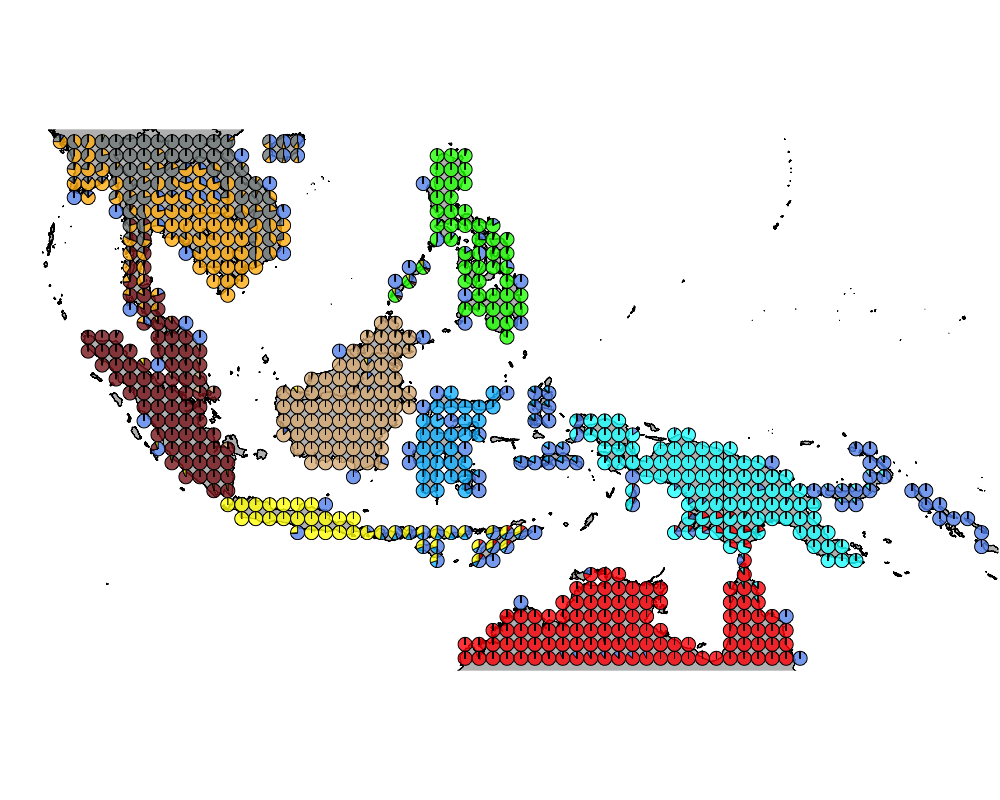
geostructure10
Key Birds
We obtain the driving bird species for each cluster using the CountClust package.
K =2
driving_species_ind <- ExtractTopFeatures(topics_clust[[2]]$freq, method = "poisson", options = "min", top_features = 50)
species_names <- apply(driving_species_ind$indices, c(1,2), function(x) return (rownames(topics_clust[[2]]$freq)[x]))
t(species_names)## [,1] [,2]
## [1,] "Rhipidura leucophrys" "Eumyias thalassinus"
## [2,] "Coracina papuensis" "Dicaeum chrysorrheum"
## [3,] "Aquila audax" "Dicrurus aeneus"
## [4,] "Chalcites basalis" "Anthreptes singalensis"
## [5,] "Elseyornis melanops" "Eurylaimus javanicus"
## [6,] "Eurostopodus argus" "Malacocincla abbotti"
## [7,] "Hamirostra melanosternon" "Ketupa ketupu"
## [8,] "Pachycephala rufiventris" "Sitta frontalis"
## [9,] "Podargus strigoides" "Chloropsis cochinchinensis"
## [10,] "Smicrornis brevirostris" "Ninox scutulata"
## [11,] "Threskiornis spinicollis" "Prinia flaviventris"
## [12,] "Todiramphus sanctus" "Prionochilus maculatus"
## [13,] "Artamus minor" "Rhinortha chlorophaea"
## [14,] "Calyptorhynchus banksii" "Meiglyptes grammithorax"
## [15,] "Chlamydera nuchalis" "Micropternus brachyurus"
## [16,] "Elanus axillaris" "Ducula badia"
## [17,] "Heteroscenes pallidus" "Hemicircus sordidus"
## [18,] "Hieraaetus morphnoides" "Strix leptogrammica"
## [19,] "Malurus melanocephalus" "Anthracoceros albirostris"
## [20,] "Taeniopygia bichenovii" "Abroscopus superciliaris"
## [21,] "Todiramphus pyrrhopygius" "Dicaeum cruentatum"
## [22,] "Ardea pacifica" "Copsychus malabaricus"
## [23,] "Aythya australis" "Tricholestes criniger"
## [24,] "Dendrocygna eytoni" "Stachyris erythroptera"
## [25,] "Geopelia cuneata" "Hemipus picatus"
## [26,] "Turnix pyrrhothorax" "Cuculus micropterus"
## [27,] "Corvus orru" "Tephrodornis gularis"
## [28,] "Certhionyx pectoralis" "Stachyris rufifrons"
## [29,] "Cracticus nigrogularis" "Phaenicophaeus diardi"
## [30,] "Conopophila rufogularis" "Phaenicophaeus sumatranus"
## [31,] "Chalcites osculans" "Corydon sumatranus"
## [32,] "Eolophus roseicapilla" "Blythipicus rubiginosus"
## [33,] "Entomyzon cyanotis" "Alcedo peninsulae"
## [34,] "Artamus personatus" "Enicurus leschenaulti"
## [35,] "Pelecanus conspicillatus" "Platysmurus leucopterus"
## [36,] "Lichenostomus unicolor" "Nyctyornis amictus"
## [37,] "Falco berigora" "Pycnonotus erythropthalmos"
## [38,] "Eudynamys orientalis" "Rollulus rouloul"
## [39,] "Grallina cyanoleuca" "Arachnothera flavigaster"
## [40,] "Cacatua galerita" "Ixos malaccensis"
## [41,] "Dicaeum hirundinaceum" "Cacomantis sonneratii"
## [42,] "Psitteuteles versicolor" "Treron capellei"
## [43,] "Haliastur sphenurus" "Iole olivacea"
## [44,] "Microeca fascinans" "Harpactes duvaucelii"
## [45,] "Ceyx azureus" "Irena puella"
## [46,] "Accipiter cirrocephalus" "Aegithina viridissima"
## [47,] "Radjah radjah" "Malacocincla malaccensis"
## [48,] "Falco cenchroides" "Aegithina tiphia"
## [49,] "Ardea plumifera" "Dicrurus paradiseus"
## [50,] "Lichenostomus virescens" "Picoides canicapillus"K = 3
driving_species_ind <- ExtractTopFeatures(topics_clust[[3]]$freq, method = "poisson", options = "min")
species_names <- apply(driving_species_ind$indices, c(1,2), function(x) return (rownames(topics_clust[[10]]$freq)[x]))
t(species_names)## [,1] [,2]
## [1,] "Coracina papuensis" "Centropus rectunguis"
## [2,] "Rhipidura leucophrys" "Melanoperdix niger"
## [3,] "Corvus orru" "Trichixos pyrropygus"
## [4,] "Cacatua galerita" "Prionochilus thoracicus"
## [5,] "Accipiter cirrocephalus" "Rhinomyias umbratilis"
## [6,] "Ardea plumifera" "Rhipidura perlata"
## [7,] "Ceyx azureus" "Lyncornis temminckii"
## [8,] "Radjah radjah" "Caprimulgus concretus"
## [9,] "Falco berigora" "Cyornis caerulatus"
## [10,] "Rhipidura rufiventris" "Ciconia stormi"
## [,3]
## [1,] "Prinia hodgsonii"
## [2,] "Sterna aurantia"
## [3,] "Chloropsis aurifrons"
## [4,] "Merops orientalis"
## [5,] "Anas poecilorhyncha"
## [6,] "Artamus fuscus"
## [7,] "Clamator coromandus"
## [8,] "Glaucidium cuculoides"
## [9,] "Coracias affinis"
## [10,] "Treron phayrei"SessionInfo
sessionInfo()## R version 3.5.0 (2018-04-23)
## Platform: x86_64-apple-darwin15.6.0 (64-bit)
## Running under: macOS Sierra 10.12.6
##
## Matrix products: default
## BLAS: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRblas.0.dylib
## LAPACK: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRlapack.dylib
##
## locale:
## [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] ggthemes_3.5.0 scales_0.5.0 mapplots_1.5
## [4] mapdata_2.3.0 maps_3.3.0 rgdal_1.2-20
## [7] gtools_3.5.0 rasterVis_0.44 latticeExtra_0.6-28
## [10] RColorBrewer_1.1-2 lattice_0.20-35 raster_2.6-7
## [13] sp_1.2-7 CountClust_1.6.1 ggplot2_2.2.1
## [16] methClust_0.1.0
##
## loaded via a namespace (and not attached):
## [1] zoo_1.8-1 modeltools_0.2-21 slam_0.1-43
## [4] reshape2_1.4.3 colorspace_1.3-2 htmltools_0.3.6
## [7] stats4_3.5.0 viridisLite_0.3.0 yaml_2.1.19
## [10] mgcv_1.8-23 rlang_0.2.0 hexbin_1.27.2
## [13] pillar_1.2.2 plyr_1.8.4 stringr_1.3.1
## [16] munsell_0.4.3 gtable_0.2.0 evaluate_0.10.1
## [19] labeling_0.3 knitr_1.20 permute_0.9-4
## [22] flexmix_2.3-14 parallel_3.5.0 Rcpp_0.12.17
## [25] backports_1.1.2 limma_3.36.1 vegan_2.5-1
## [28] maptpx_1.9-5 picante_1.7 digest_0.6.15
## [31] stringi_1.2.2 cowplot_0.9.2 grid_3.5.0
## [34] rprojroot_1.3-2 tools_3.5.0 magrittr_1.5
## [37] lazyeval_0.2.1 tibble_1.4.2 cluster_2.0.7-1
## [40] ape_5.1 MASS_7.3-49 Matrix_1.2-14
## [43] SQUAREM_2017.10-1 assertthat_0.2.0 rmarkdown_1.9
## [46] boot_1.3-20 nnet_7.3-12 nlme_3.1-137
## [49] compiler_3.5.0This R Markdown site was created with workflowr