Intro
Here we look at those regions in Wallacea which has high admixture in terms of the presence of distinct communities of mammals.
Packages
library(methClust)
library(CountClust)
library(rasterVis)
library(gtools)
library(sp)
library(rgdal)
library(ggplot2)
library(maps)
library(mapdata)
library(mapplots)
library(scales)
library(ggthemes)Load the data
Wallacea Region data
mamms <- get(load("../data/mammals_with_bats.rda"))
latlong_chars <- rownames(mamms)
latlong <- cbind.data.frame(as.numeric(sapply(latlong_chars,
function(x) return(strsplit(x, "_")[[1]][1]))),
as.numeric(sapply(latlong_chars,
function(x) return(strsplit(x, "_")[[1]][2]))))GoM model
topics_clust <- get(load("../output/methClust_wallacea_mammals_bats.rda"))K = 8
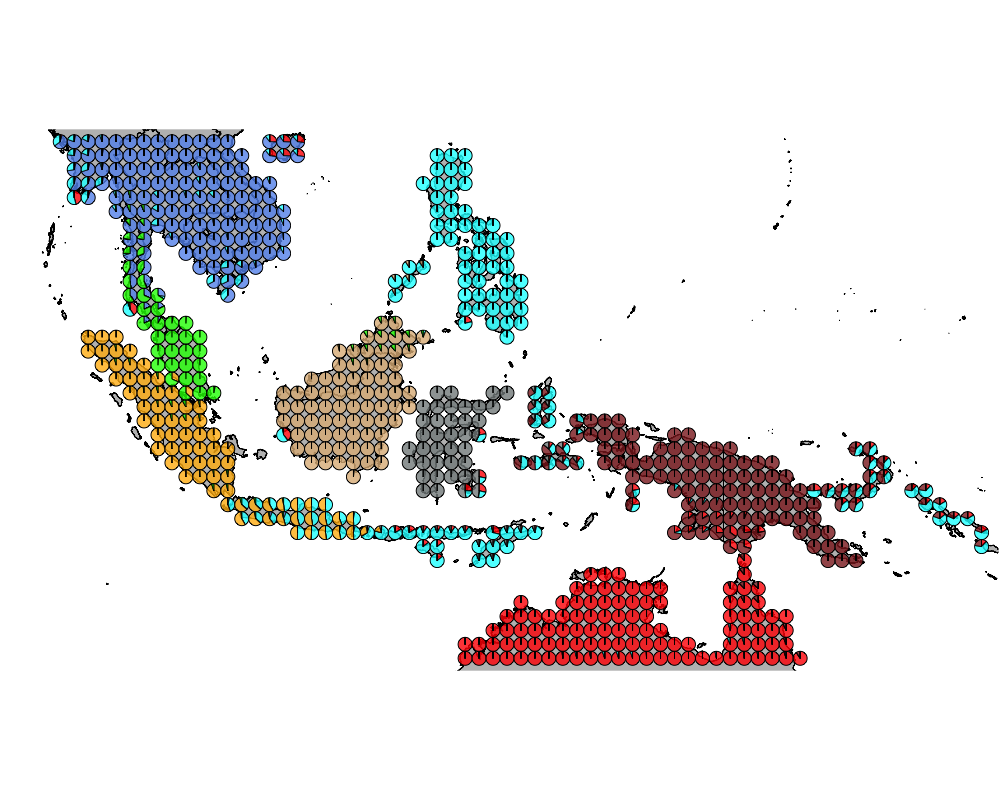
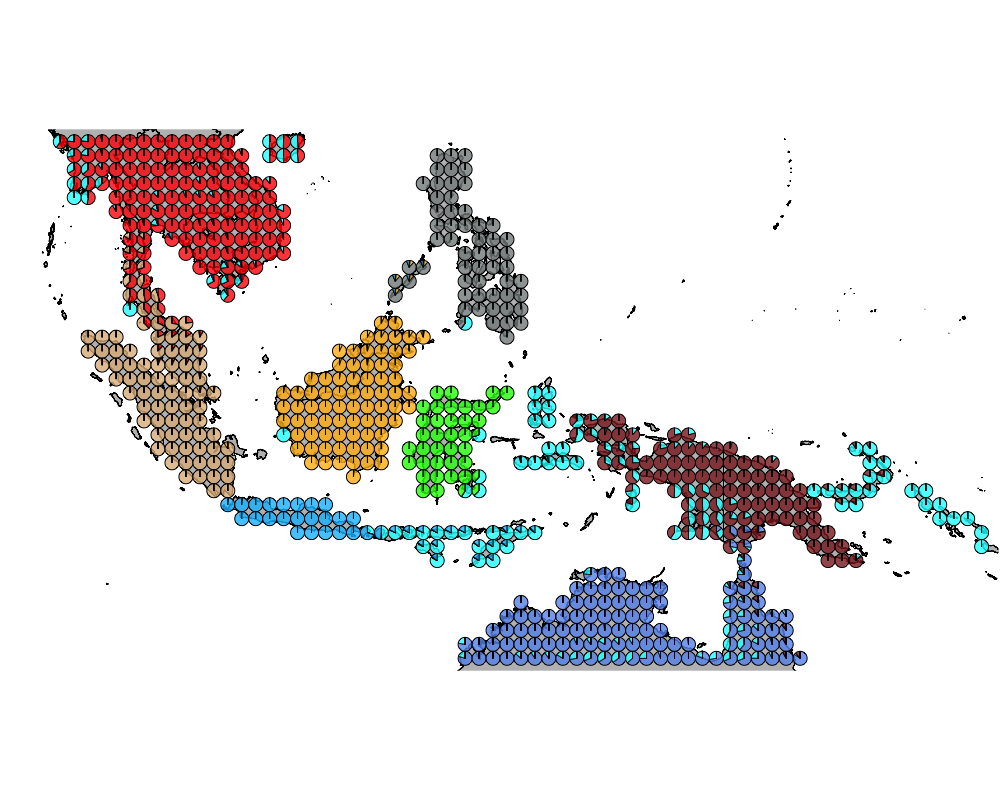
K = 9
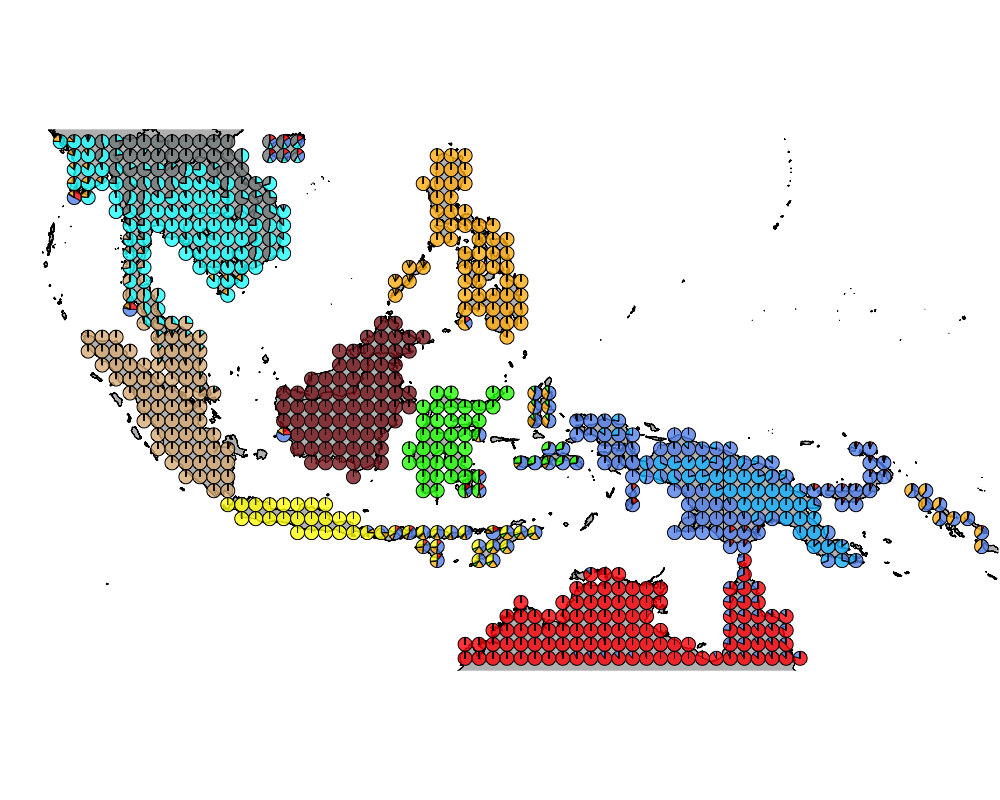
K = 10
Identifying number of mixed components
K = 8
topics <- topics_clust[[8]]
num_components <- apply(topics$omega, 1, function(x) return (length(which(x > 0.1))))
modelmat <- model.matrix(~as.factor(num_components)-1)color = colorRampPalette(c("white", "red"))(5)
intensity <- 0.8
for(m in 1:1){
png(filename=paste0("../docs/High_ADMIX_regions_mammals/high_admixture_K_8.png"),width = 1000, height = 800)
map("worldHires",
ylim=c(-18,20), xlim=c(90,160), # Re-defines the latitude and longitude range
col = "gray", fill=TRUE, mar=c(0.1,0.1,0.1,0.1))
lapply(1:dim(modelmat)[1], function(r)
add.pie(z=as.integer(100*modelmat[r,]),
x=latlong[r,1], y=latlong[r,2], labels=c("","",""),
radius = 0.5,
col=c(alpha(color[1],intensity),alpha(color[2],intensity),
alpha(color[3], intensity), alpha(color[4], intensity),
alpha(color[5], intensity))));
dev.off()
}K = 9
topics <- topics_clust[[9]]
num_components <- apply(topics$omega, 1, function(x) return (length(which(x > 0.1))))
modelmat <- model.matrix(~as.factor(num_components)-1)color = colorRampPalette(c("white", "red"))(5)
intensity <- 0.8
for(m in 1:1){
png(filename=paste0("../docs/High_ADMIX_regions_mammals/high_admixture_K_9.png"),width = 1000, height = 800)
map("worldHires",
ylim=c(-18,20), xlim=c(90,160), # Re-defines the latitude and longitude range
col = "gray", fill=TRUE, mar=c(0.1,0.1,0.1,0.1))
lapply(1:dim(modelmat)[1], function(r)
add.pie(z=as.integer(100*modelmat[r,]),
x=latlong[r,1], y=latlong[r,2], labels=c("","",""),
radius = 0.5,
col=c(alpha(color[1],intensity),alpha(color[2],intensity),
alpha(color[3], intensity), alpha(color[4], intensity),
alpha(color[5], intensity))));
dev.off()
}K = 10
topics <- topics_clust[[10]]
num_components <- apply(topics$omega, 1, function(x) return (length(which(x > 0.1))))
modelmat <- model.matrix(~as.factor(num_components)-1)color = colorRampPalette(c("white", "red"))(5)
intensity <- 0.8
for(m in 1:1){
png(filename=paste0("../docs/High_ADMIX_regions_mammals/high_admixture_K_10.png"),width = 1000, height = 800)
map("worldHires",
ylim=c(-18,20), xlim=c(90,160), # Re-defines the latitude and longitude range
col = "gray", fill=TRUE, mar=c(0.1,0.1,0.1,0.1))
lapply(1:dim(modelmat)[1], function(r)
add.pie(z=as.integer(100*modelmat[r,]),
x=latlong[r,1], y=latlong[r,2], labels=c("","",""),
radius = 0.5,
col=c(alpha(color[1],intensity),alpha(color[2],intensity),
alpha(color[3], intensity), alpha(color[4], intensity),
alpha(color[5], intensity))));
dev.off()
}Visualization of high admix regions
K = 8
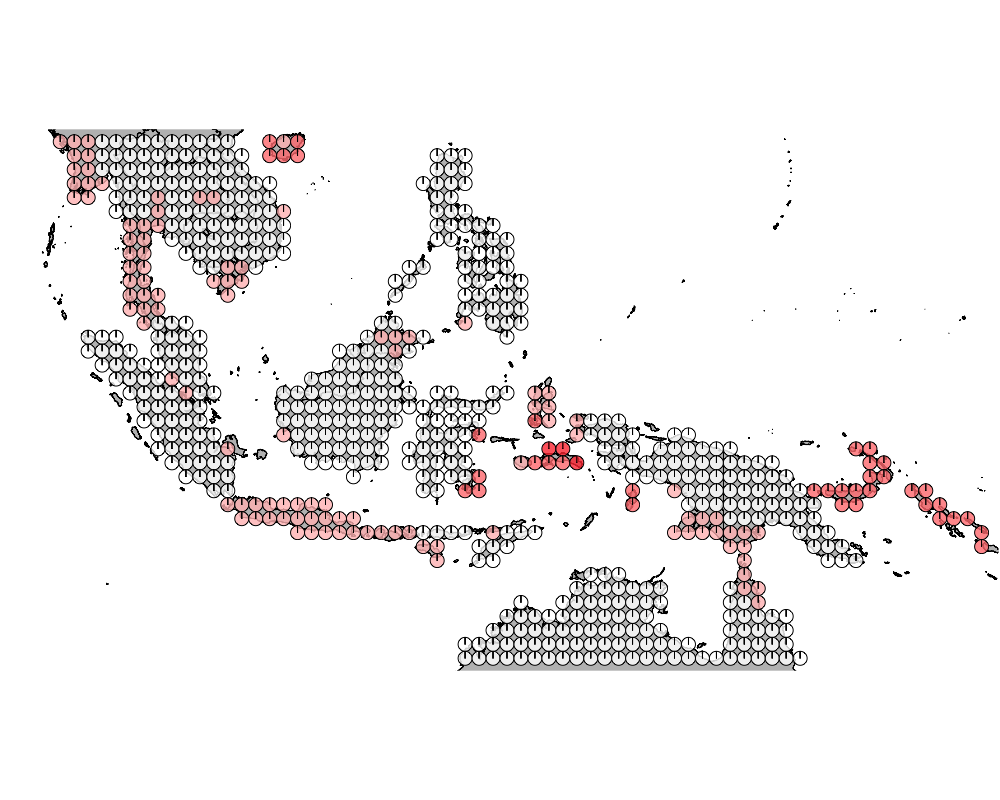
K = 9
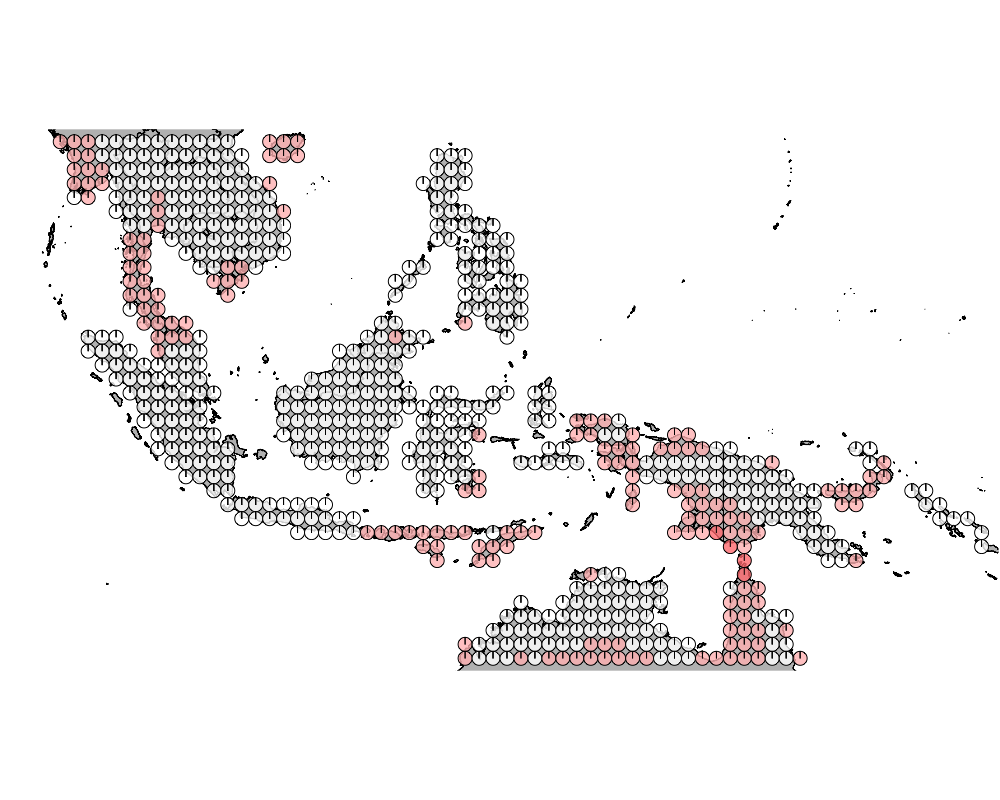
geostructure10
K = 10
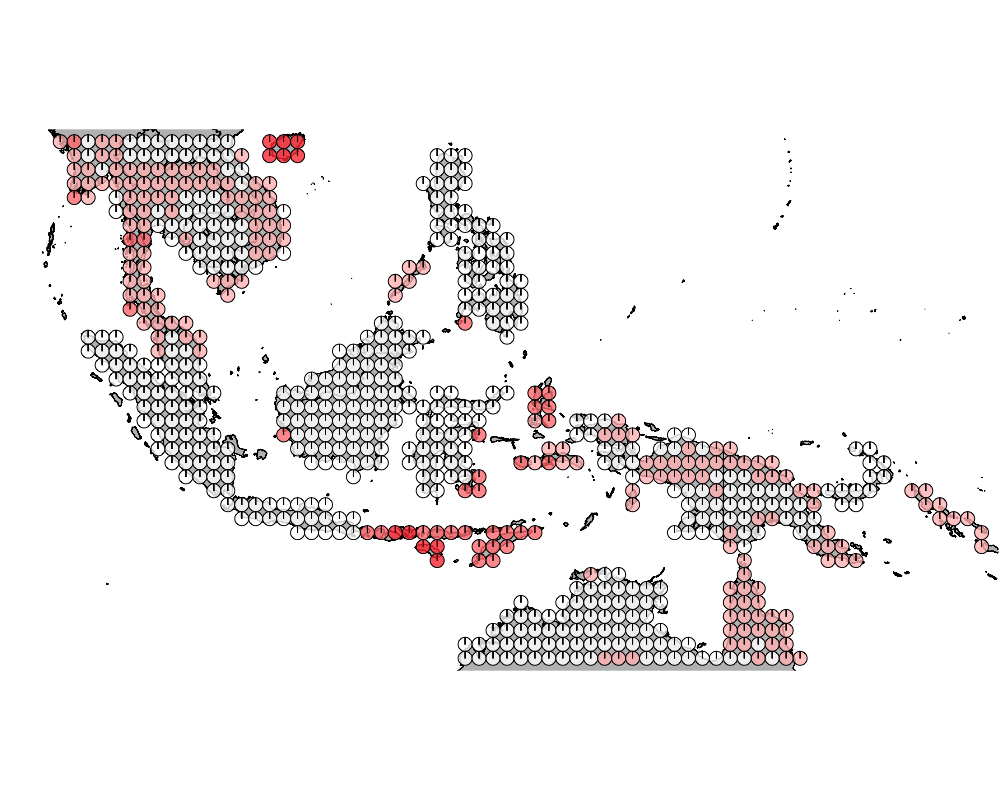
geostructure10
sessionInfo()## R version 3.5.0 (2018-04-23)
## Platform: x86_64-apple-darwin15.6.0 (64-bit)
## Running under: macOS Sierra 10.12.6
##
## Matrix products: default
## BLAS: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRblas.0.dylib
## LAPACK: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRlapack.dylib
##
## locale:
## [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] ggthemes_3.5.0 scales_0.5.0 mapplots_1.5
## [4] mapdata_2.3.0 maps_3.3.0 rgdal_1.2-20
## [7] gtools_3.5.0 rasterVis_0.44 latticeExtra_0.6-28
## [10] RColorBrewer_1.1-2 lattice_0.20-35 raster_2.6-7
## [13] sp_1.2-7 CountClust_1.6.1 ggplot2_2.2.1
## [16] methClust_0.1.0
##
## loaded via a namespace (and not attached):
## [1] zoo_1.8-1 modeltools_0.2-21 slam_0.1-43
## [4] reshape2_1.4.3 colorspace_1.3-2 htmltools_0.3.6
## [7] stats4_3.5.0 viridisLite_0.3.0 yaml_2.1.19
## [10] mgcv_1.8-23 rlang_0.2.0 hexbin_1.27.2
## [13] pillar_1.2.2 plyr_1.8.4 stringr_1.3.1
## [16] munsell_0.4.3 gtable_0.2.0 evaluate_0.10.1
## [19] knitr_1.20 permute_0.9-4 flexmix_2.3-14
## [22] parallel_3.5.0 Rcpp_0.12.17 backports_1.1.2
## [25] limma_3.36.1 vegan_2.5-1 maptpx_1.9-5
## [28] picante_1.7 digest_0.6.15 stringi_1.2.2
## [31] cowplot_0.9.2 grid_3.5.0 rprojroot_1.3-2
## [34] tools_3.5.0 magrittr_1.5 lazyeval_0.2.1
## [37] tibble_1.4.2 cluster_2.0.7-1 ape_5.1
## [40] MASS_7.3-49 Matrix_1.2-14 SQUAREM_2017.10-1
## [43] assertthat_0.2.0 rmarkdown_1.9 boot_1.3-20
## [46] nnet_7.3-12 nlme_3.1-137 compiler_3.5.0This R Markdown site was created with workflowr