High admixture regions in the Wallace region
Kushal K Dey
4/12/2018
Introduction
In this script, we look at the high admixture regions in Wallacea. High admixture is defined as having substantial memberships in many clusters. Usually each grid cell has substantial membership (say > 0.1) in one cluster - but some grid cells may have an admixing of the different communities of bird species and they will show high memberships in multiple clusters.
Packages
library(methClust)
library(CountClust)
library(rasterVis)
library(gtools)
library(sp)
library(rgdal)
library(ggplot2)
library(maps)
library(mapdata)
library(mapplots)
library(scales)
library(ggthemes)Load the data
Wallacea Region data
birds <- get(load("../data/wallacea_birds.rda"))
latlong_chars_birds <- rownames(birds)
latlong <- cbind.data.frame(as.numeric(sapply(latlong_chars_birds,
function(x) return(strsplit(x, "_")[[1]][1]))),
as.numeric(sapply(latlong_chars_birds,
function(x) return(strsplit(x, "_")[[1]][2]))))GoM model
topics_clust <- get(load("../output/methClust_wallacea_birds.rda"))ecoSTRUCTURE plot visualization
K = 8
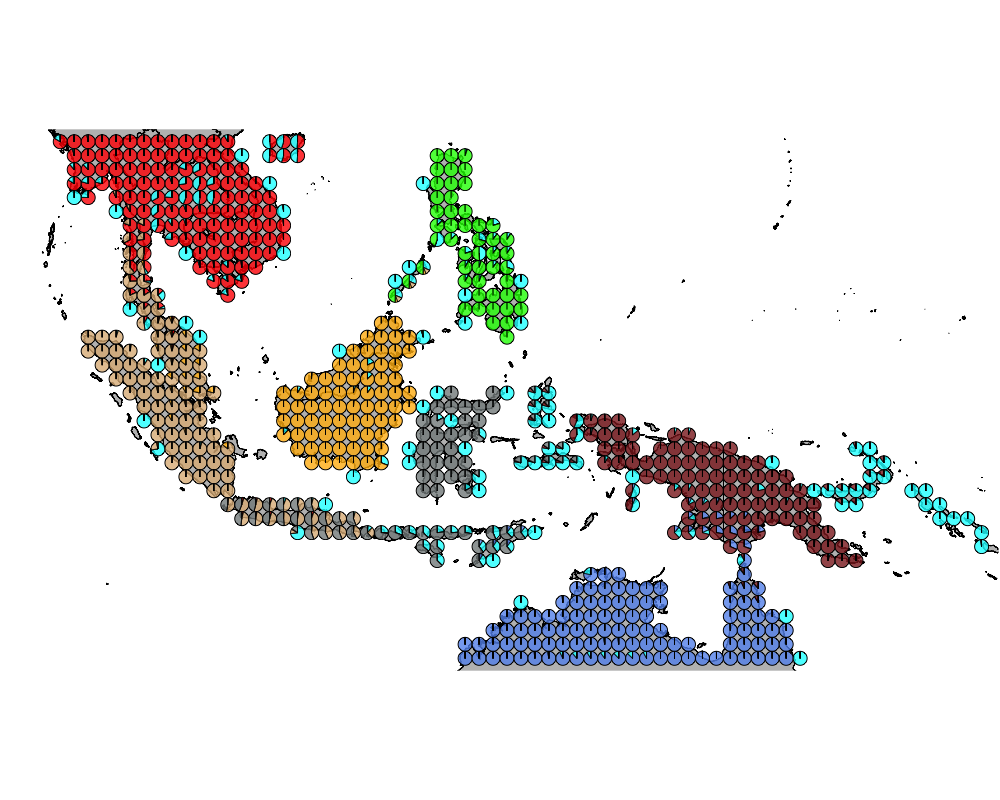
geostructure10
K = 9
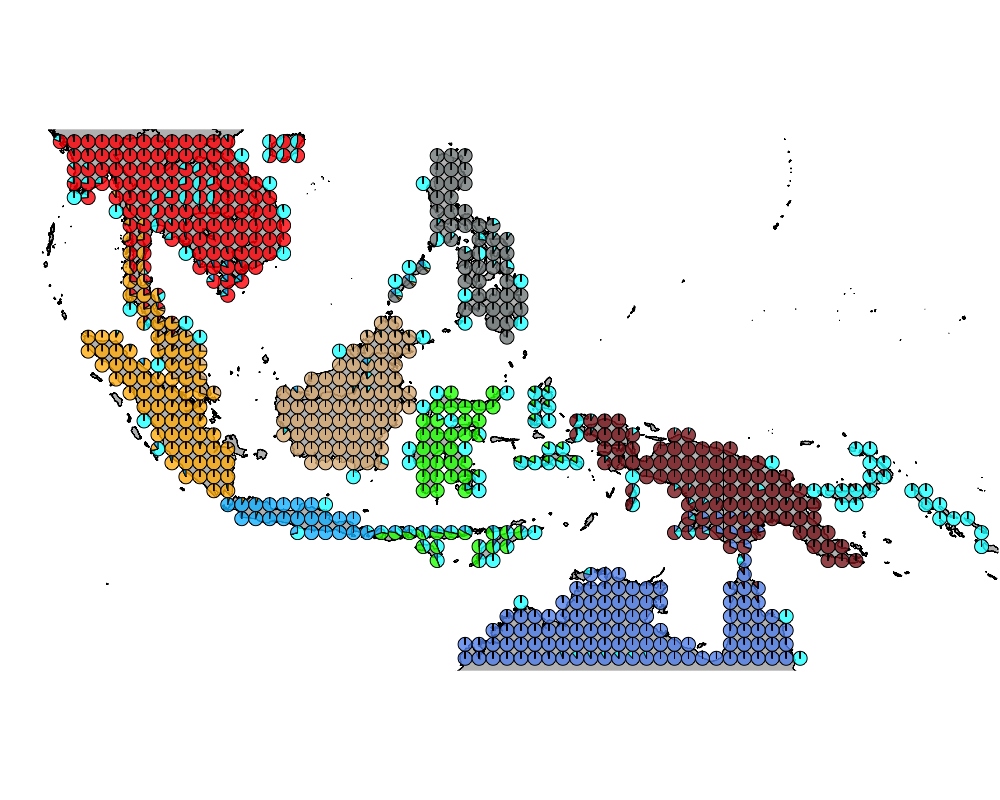
geostructure10
K = 10
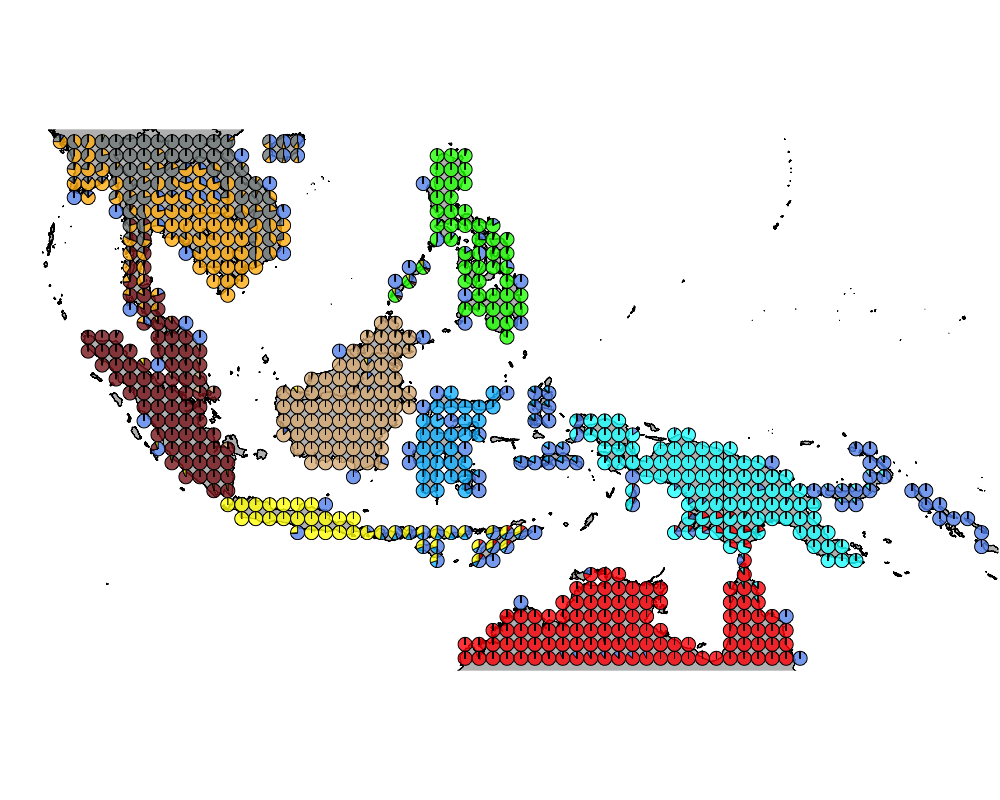
geostructure10
Identifying number of mixed components
K = 8
topics <- topics_clust[[8]]
num_components <- apply(topics$omega, 1, function(x) return (length(which(x > 0.1))))
modelmat <- model.matrix(~as.factor(num_components)-1)color = colorRampPalette(c("white", "red"))(5)
intensity <- 0.8
for(m in 1:1){
png(filename=paste0("../docs/High_ADMIX_regions_birds/high_admixture_K_8.png"),width = 1000, height = 800)
map("worldHires",
ylim=c(-18,20), xlim=c(90,160), # Re-defines the latitude and longitude range
col = "gray", fill=TRUE, mar=c(0.1,0.1,0.1,0.1))
lapply(1:dim(modelmat)[1], function(r)
add.pie(z=as.integer(100*modelmat[r,]),
x=latlong[r,1], y=latlong[r,2], labels=c("","",""),
radius = 0.5,
col=c(alpha(color[1],intensity),alpha(color[2],intensity),
alpha(color[3], intensity), alpha(color[4], intensity),
alpha(color[5], intensity))));
dev.off()
}K = 9
topics <- topics_clust[[9]]
num_components <- apply(topics$omega, 1, function(x) return (length(which(x > 0.1))))
modelmat <- model.matrix(~as.factor(num_components)-1)color = colorRampPalette(c("white", "red"))(5)
intensity <- 0.8
for(m in 1:1){
png(filename=paste0("../docs/High_ADMIX_regions_birds/high_admixture_K_9.png"),width = 1000, height = 800)
map("worldHires",
ylim=c(-18,20), xlim=c(90,160), # Re-defines the latitude and longitude range
col = "gray", fill=TRUE, mar=c(0.1,0.1,0.1,0.1))
lapply(1:dim(modelmat)[1], function(r)
add.pie(z=as.integer(100*modelmat[r,]),
x=latlong[r,1], y=latlong[r,2], labels=c("","",""),
radius = 0.5,
col=c(alpha(color[1],intensity),alpha(color[2],intensity),
alpha(color[3], intensity), alpha(color[4], intensity),
alpha(color[5], intensity))));
dev.off()
}K = 10
topics <- topics_clust[[10]]
num_components <- apply(topics$omega, 1, function(x) return (length(which(x > 0.1))))
modelmat <- model.matrix(~as.factor(num_components)-1)color = colorRampPalette(c("white", "red"))(5)
intensity <- 0.8
for(m in 1:1){
png(filename=paste0("../docs/High_ADMIX_regions_birds/high_admixture_K_10.png"),width = 1000, height = 800)
map("worldHires",
ylim=c(-18,20), xlim=c(90,160), # Re-defines the latitude and longitude range
col = "gray", fill=TRUE, mar=c(0.1,0.1,0.1,0.1))
lapply(1:dim(modelmat)[1], function(r)
add.pie(z=as.integer(100*modelmat[r,]),
x=latlong[r,1], y=latlong[r,2], labels=c("","",""),
radius = 0.5,
col=c(alpha(color[1],intensity),alpha(color[2],intensity),
alpha(color[3], intensity), alpha(color[4], intensity),
alpha(color[5], intensity))));
dev.off()
}Visualization of high admix regions
K = 8
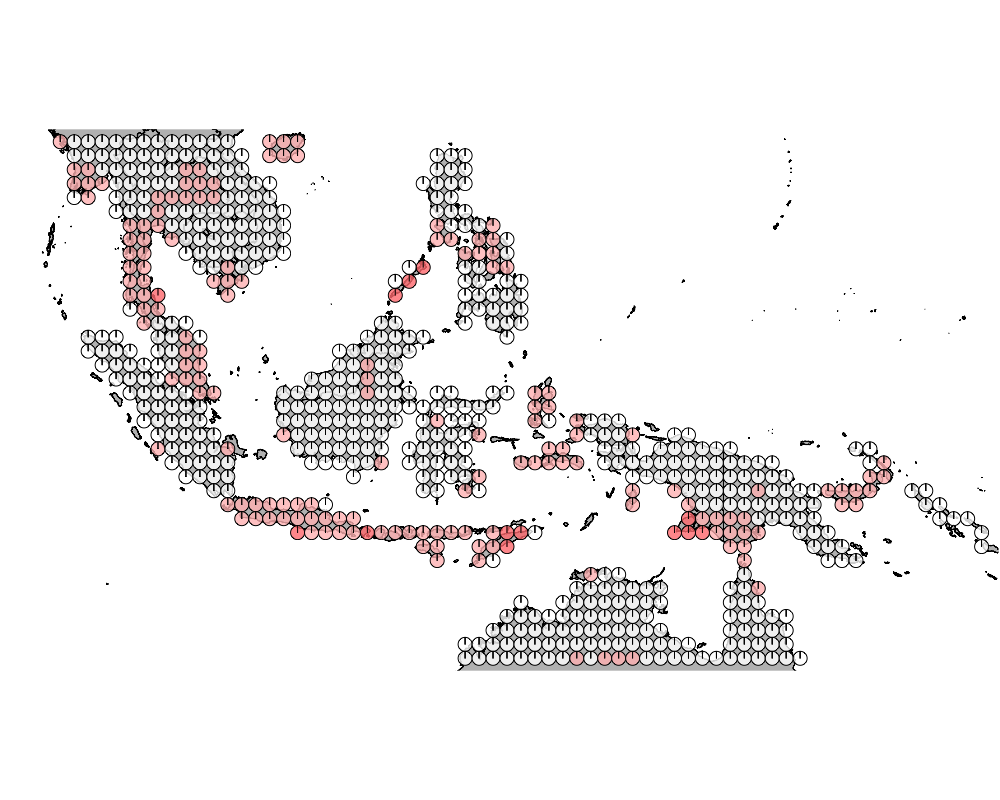
geostructure10
K = 9
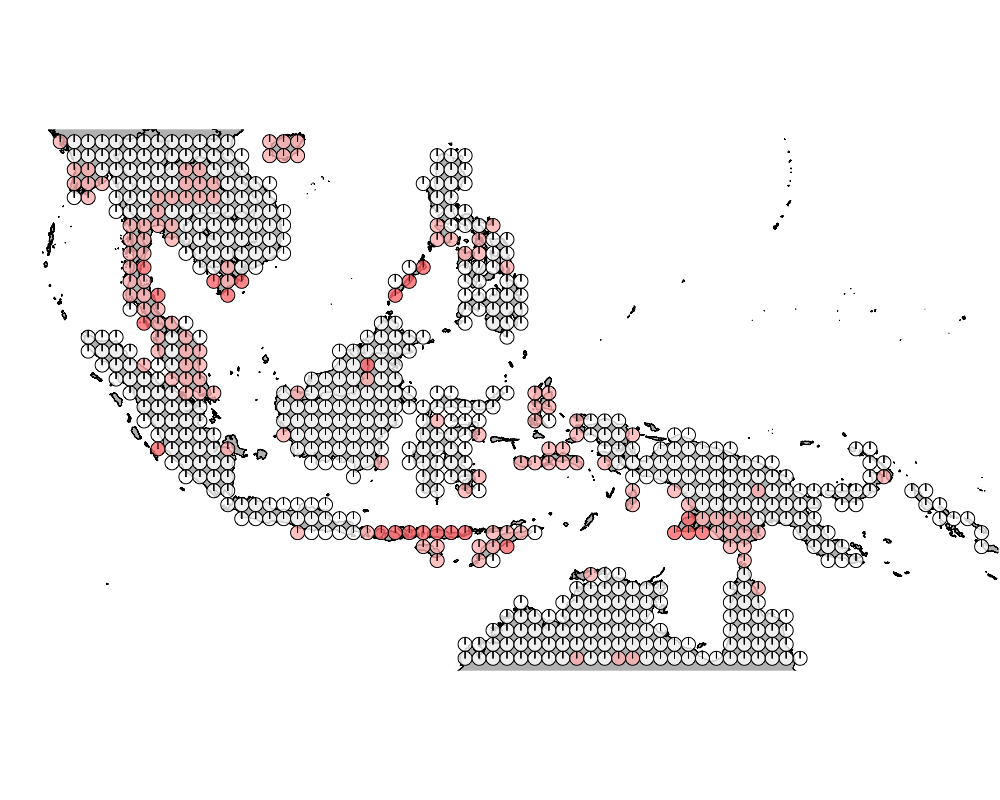
geostructure10
K = 10
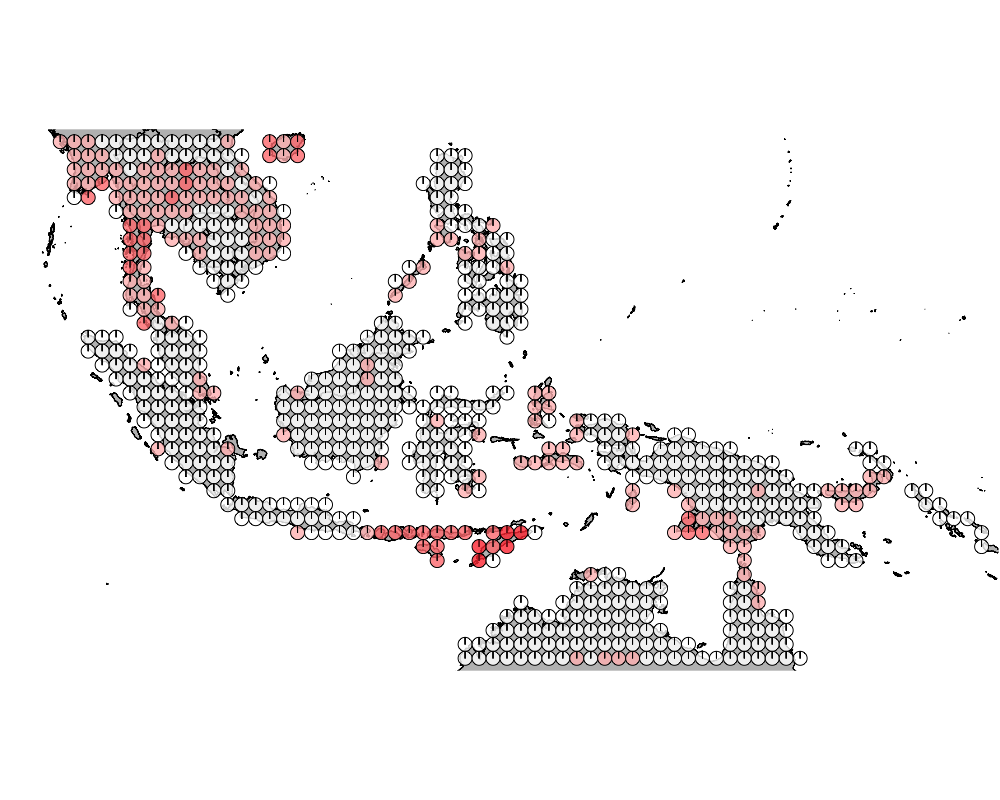
geostructure10
Overall we see that the strips of islnads in the Wallacea region are highly admixed.
SessionInfo
sessionInfo()## R version 3.5.0 (2018-04-23)
## Platform: x86_64-apple-darwin15.6.0 (64-bit)
## Running under: macOS Sierra 10.12.6
##
## Matrix products: default
## BLAS: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRblas.0.dylib
## LAPACK: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRlapack.dylib
##
## locale:
## [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] ggthemes_3.5.0 scales_0.5.0 mapplots_1.5
## [4] mapdata_2.3.0 maps_3.3.0 rgdal_1.2-20
## [7] gtools_3.5.0 rasterVis_0.44 latticeExtra_0.6-28
## [10] RColorBrewer_1.1-2 lattice_0.20-35 raster_2.6-7
## [13] sp_1.2-7 CountClust_1.6.1 ggplot2_2.2.1
## [16] methClust_0.1.0
##
## loaded via a namespace (and not attached):
## [1] zoo_1.8-1 modeltools_0.2-21 slam_0.1-43
## [4] reshape2_1.4.3 colorspace_1.3-2 htmltools_0.3.6
## [7] stats4_3.5.0 viridisLite_0.3.0 yaml_2.1.19
## [10] mgcv_1.8-23 rlang_0.2.0 hexbin_1.27.2
## [13] pillar_1.2.2 plyr_1.8.4 stringr_1.3.1
## [16] munsell_0.4.3 gtable_0.2.0 evaluate_0.10.1
## [19] knitr_1.20 permute_0.9-4 flexmix_2.3-14
## [22] parallel_3.5.0 Rcpp_0.12.17 backports_1.1.2
## [25] limma_3.36.1 vegan_2.5-1 maptpx_1.9-5
## [28] picante_1.7 digest_0.6.15 stringi_1.2.2
## [31] cowplot_0.9.2 grid_3.5.0 rprojroot_1.3-2
## [34] tools_3.5.0 magrittr_1.5 lazyeval_0.2.1
## [37] tibble_1.4.2 cluster_2.0.7-1 ape_5.1
## [40] MASS_7.3-49 Matrix_1.2-14 SQUAREM_2017.10-1
## [43] assertthat_0.2.0 rmarkdown_1.9 boot_1.3-20
## [46] nnet_7.3-12 nlme_3.1-137 compiler_3.5.0This R Markdown site was created with workflowr