Wallace Line Driving Birds
Kushal K Dey
1/2/2018
Data Processing + driving birds extraction
latlong <- get(load("../data/LatLongCells_frame.rda"))world_map <- map_data("world")
world_map <- world_map[world_map$region != "Antarctica",] # intercourse antarctica
world_map <- world_map[world_map$long > 90 & world_map$long < 160, ]
world_map <- world_map[world_map$lat > -18 & world_map$lat < 20, ]
p <- ggplot() + coord_fixed() +
xlab("") + ylab("")
#Add map to base plot
base_world_messy <- p + geom_polygon(data=world_map, aes(x=long, y=lat, group=group), colour="light green", fill="light green")
cleanup <-
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
panel.background = element_rect(fill = 'white', colour = 'white'),
axis.line = element_line(colour = "white"), legend.position="none",
axis.ticks=element_blank(), axis.text.x=element_blank(),
axis.text.y=element_blank())
base_world <- base_world_messy + cleanup
base_world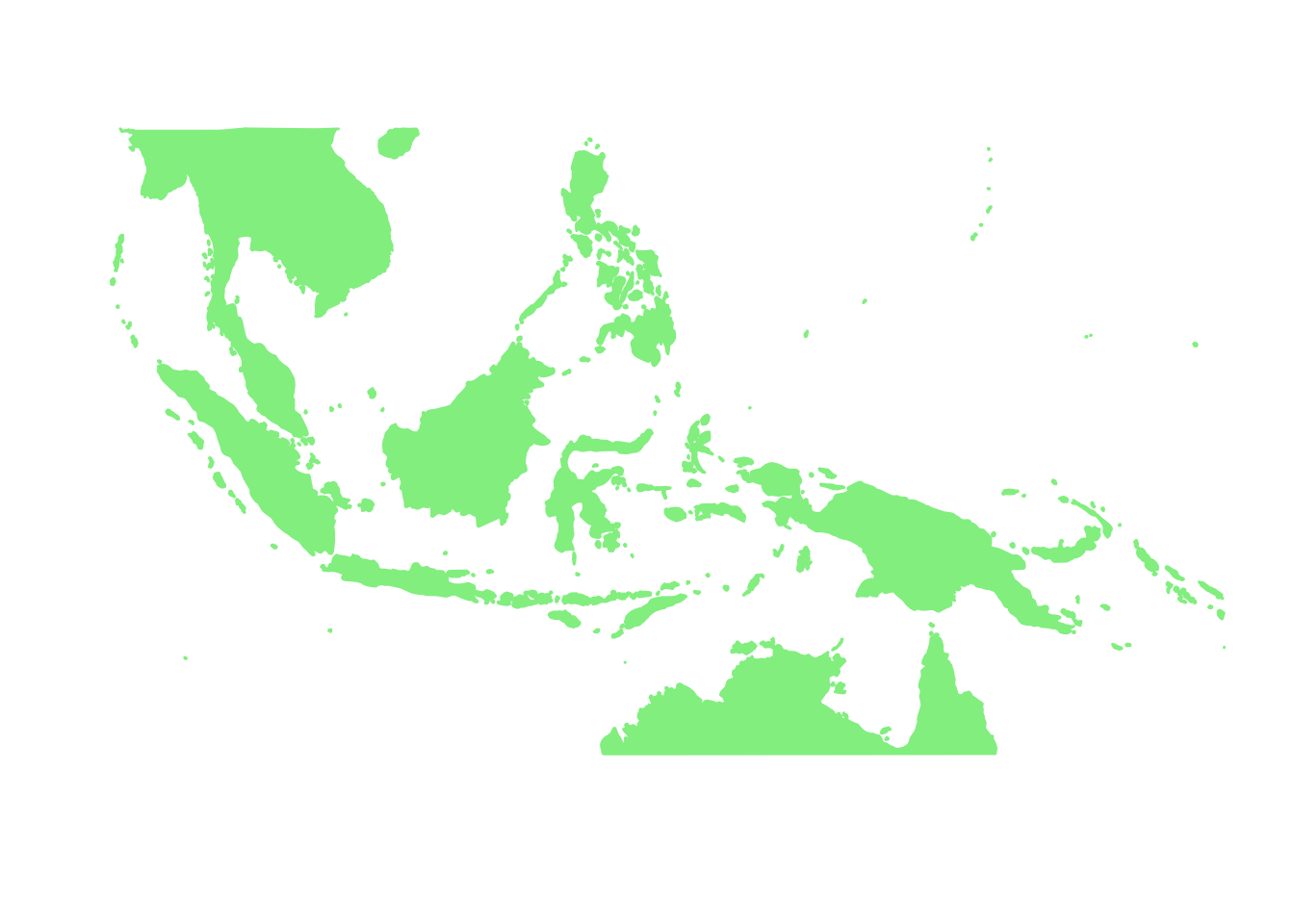
latlong <- get(load("../data/LatLongCells_frame.rda"))idx1 <- which(latlong[,2] > -18 & latlong[,2] < 20)
idx2 <- which(latlong[,1] > 90 & latlong[,1] < 160)
idx <- intersect(idx1, idx2)
length(idx)## [1] 703latlong2 <- latlong[idx,]birds_pa_data <- readRDS("../data/birds_presab_land_breeding_counts.rds")
birds_pa_data_2 <- birds_pa_data[idx, ]
birds_pa_data_3 <- birds_pa_data_2[, which(colSums(birds_pa_data_2)!=0)]Names of driving bird species
topics <- get(load("../output/Wallacea/methClust_2.rda"))second_topic_scores <- topics$freq[,2] - topics$freq[,1]
first_topic_scores <- topics$freq[,1] - topics$freq[,2]Second topic birds
names(second_topic_scores)[order(second_topic_scores, decreasing = TRUE)[1:10]]## [1] "Amaurornis phoenicurus" "Centropus bengalensis"
## [3] "Cypsiurus balasiensis" "Gallinula chloropus"
## [5] "Hypothymis azurea" "Eudynamys scolopaceus"
## [7] "Spilopelia chinensis" "Aegithina tiphia"
## [9] "Anthracoceros albirostris" "Irena puella"First topic birds
names(first_topic_scores)[order(first_topic_scores, decreasing = TRUE)[1:10]]## [1] "Anas gracilis" "Eudynamys orientalis"
## [3] "Nycticorax caledonicus" "Anas superciliosa"
## [5] "Rhipidura leucophrys" "Coracina papuensis"
## [7] "Corvus orru" "Megalurus timoriensis"
## [9] "Rhipidura rufiventris" "Cacatua galerita"Patterns of presence absence of driving bird species
PlotAssemblageIdx <- function(idx){
dat <- cbind.data.frame(latlong2, birds_pa_data_3[,idx])
colnames(dat) <- c("Latitude", "Longitude", "Value")
map_data_coloured <-
base_world +
geom_point(data=dat,
aes(x=Latitude, y=Longitude, colour=Value), size=0.5) +
scale_colour_gradient(low = "white", high = "black")
map_data_coloured
}par(mfrow = c(5,2))
for(m in 1:10){
PlotAssemblageIdx(order(second_topic_scores, decreasing = TRUE)[m])
}par(mfrow = c(5,2))
PlotAssemblageIdx(order(second_topic_scores, decreasing = TRUE)[1])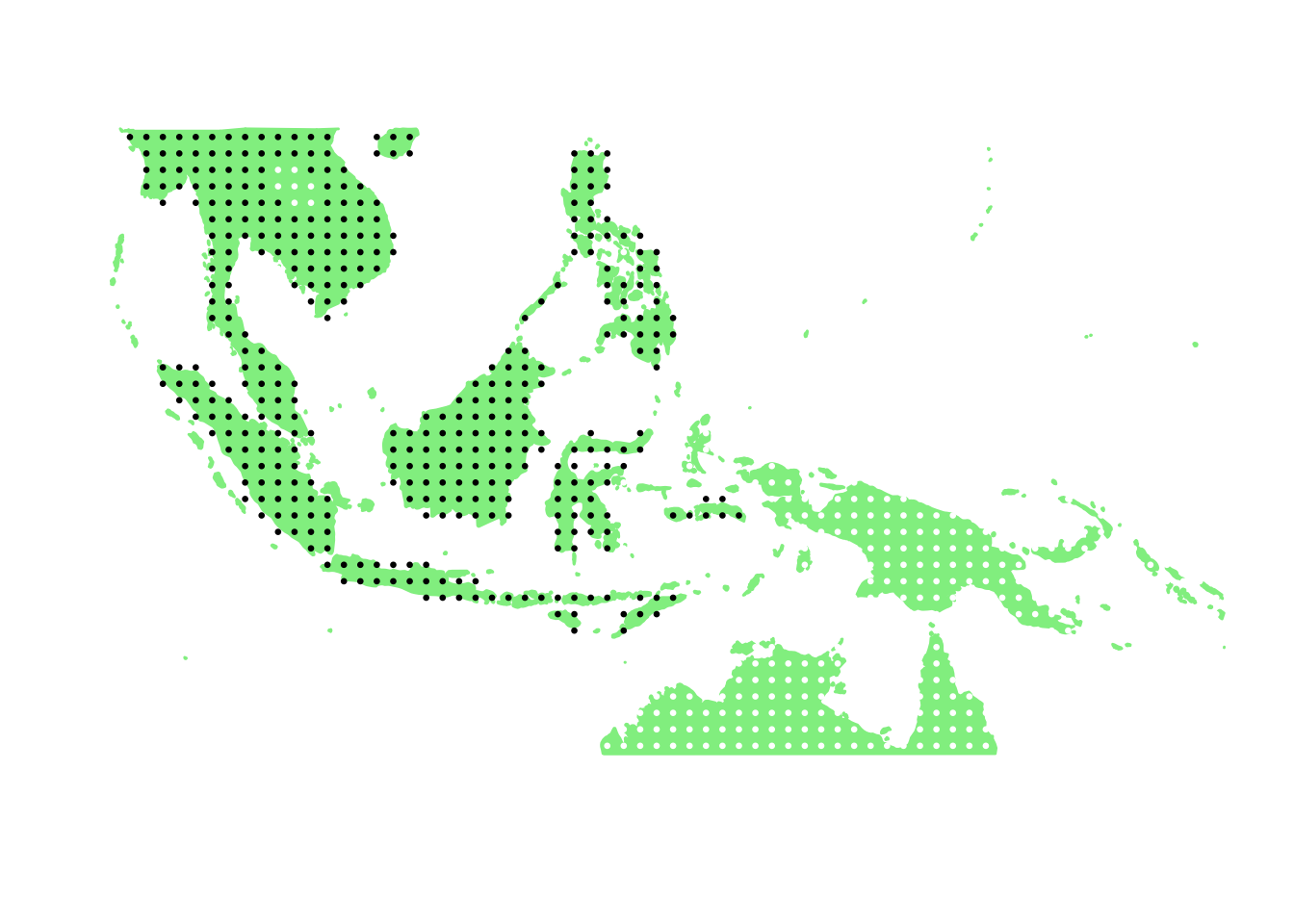
PlotAssemblageIdx(order(second_topic_scores, decreasing = TRUE)[2])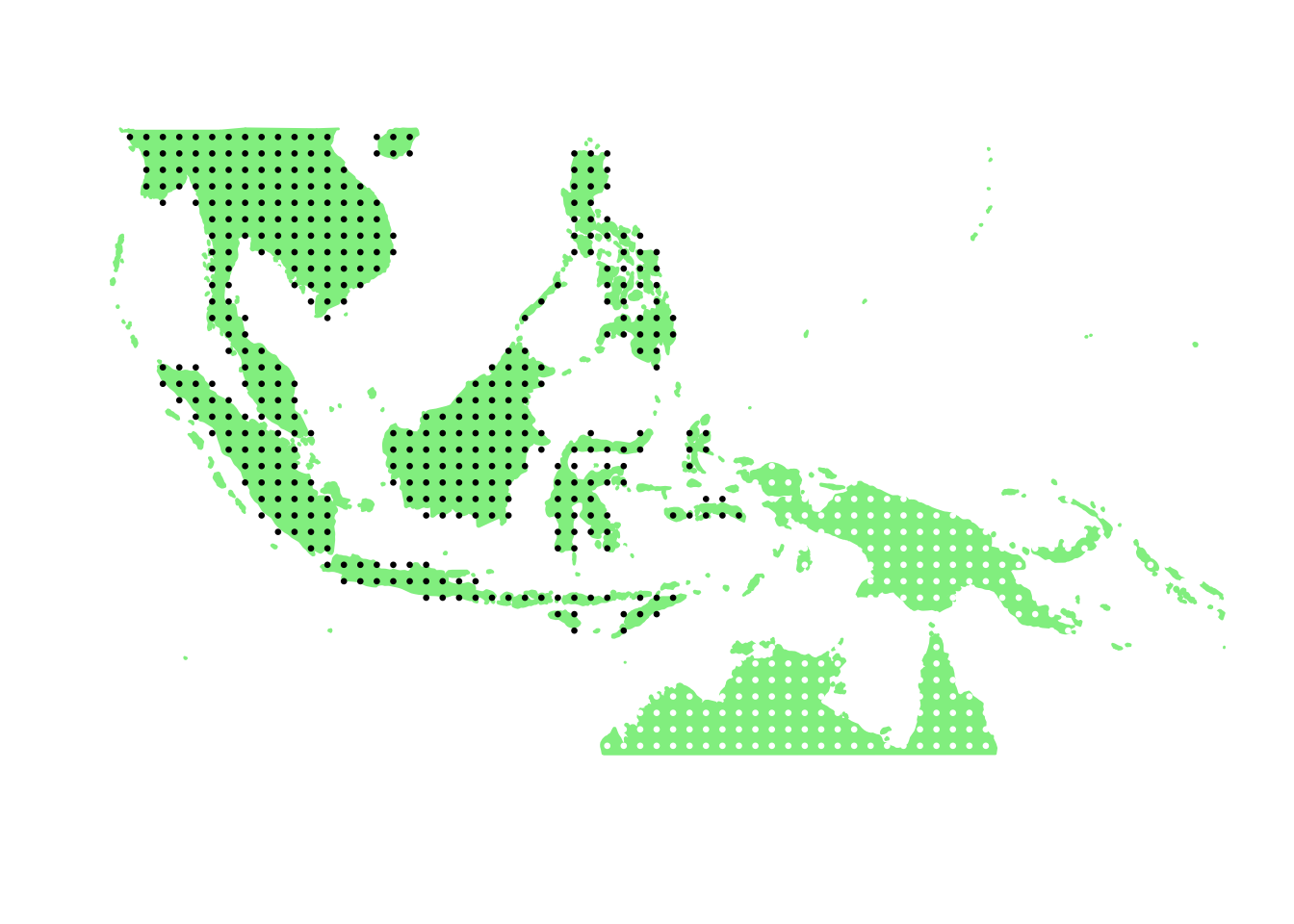
PlotAssemblageIdx(order(second_topic_scores, decreasing = TRUE)[3])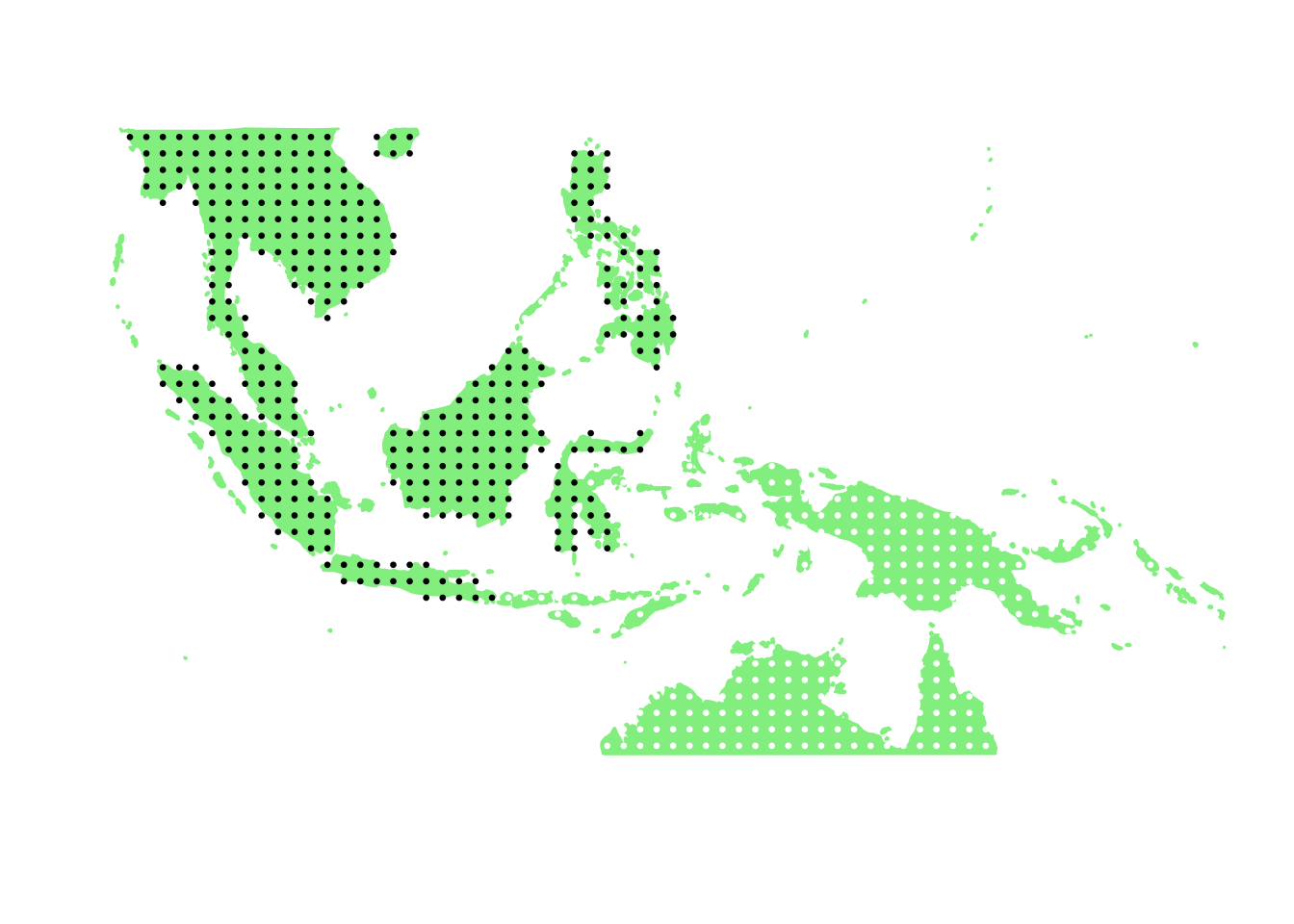
PlotAssemblageIdx(order(second_topic_scores, decreasing = TRUE)[4])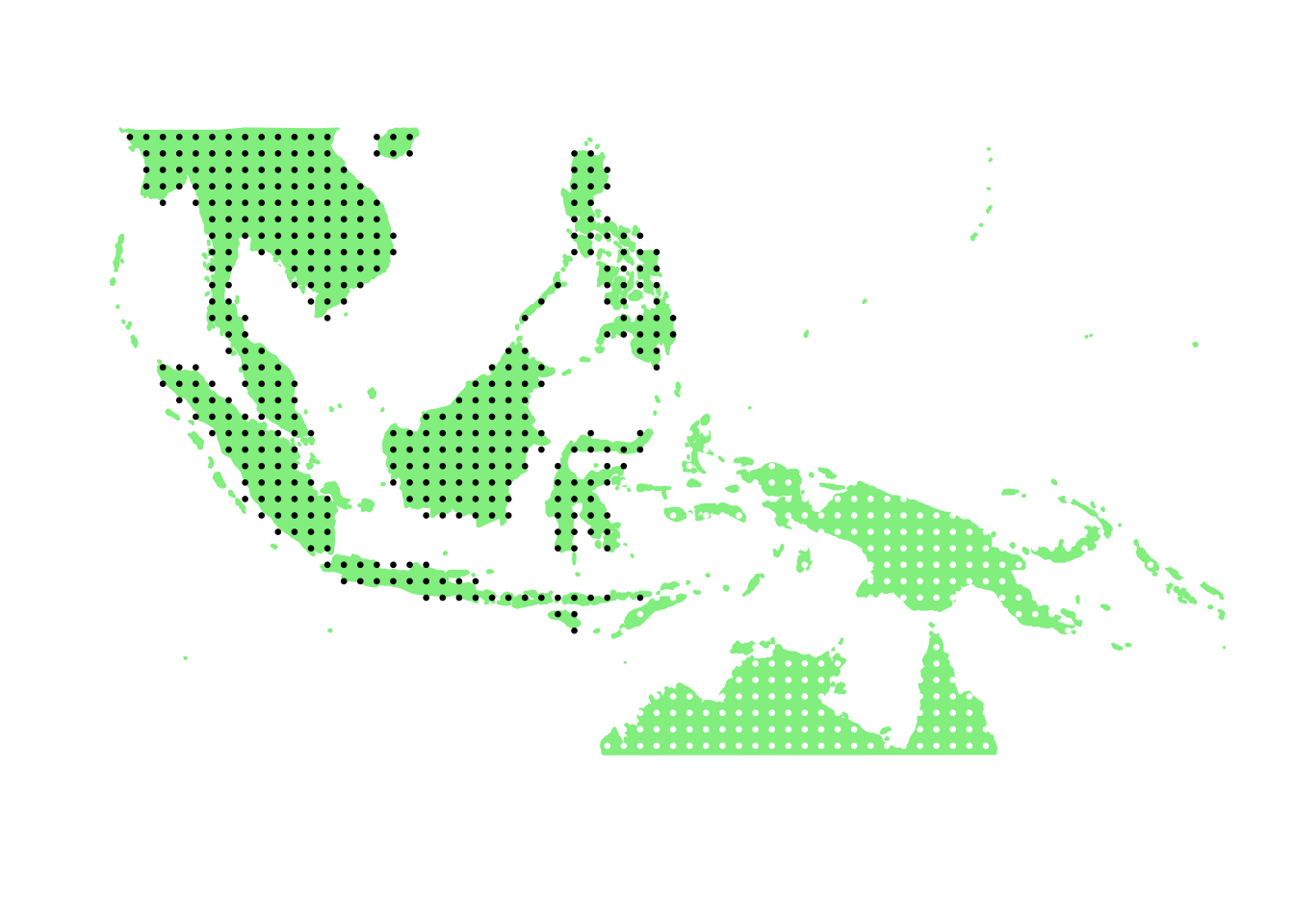
PlotAssemblageIdx(order(second_topic_scores, decreasing = TRUE)[5])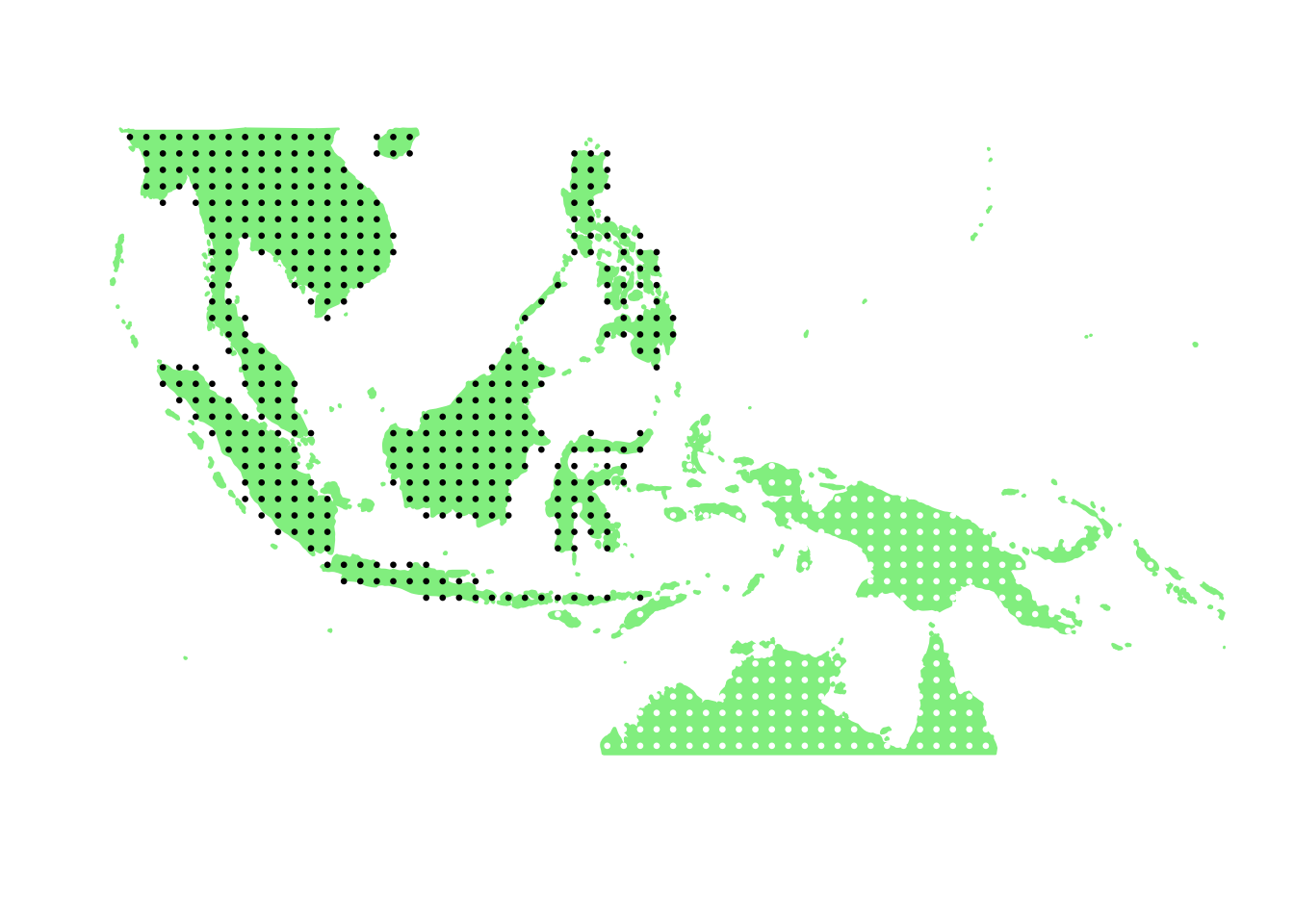
PlotAssemblageIdx(order(second_topic_scores, decreasing = TRUE)[6])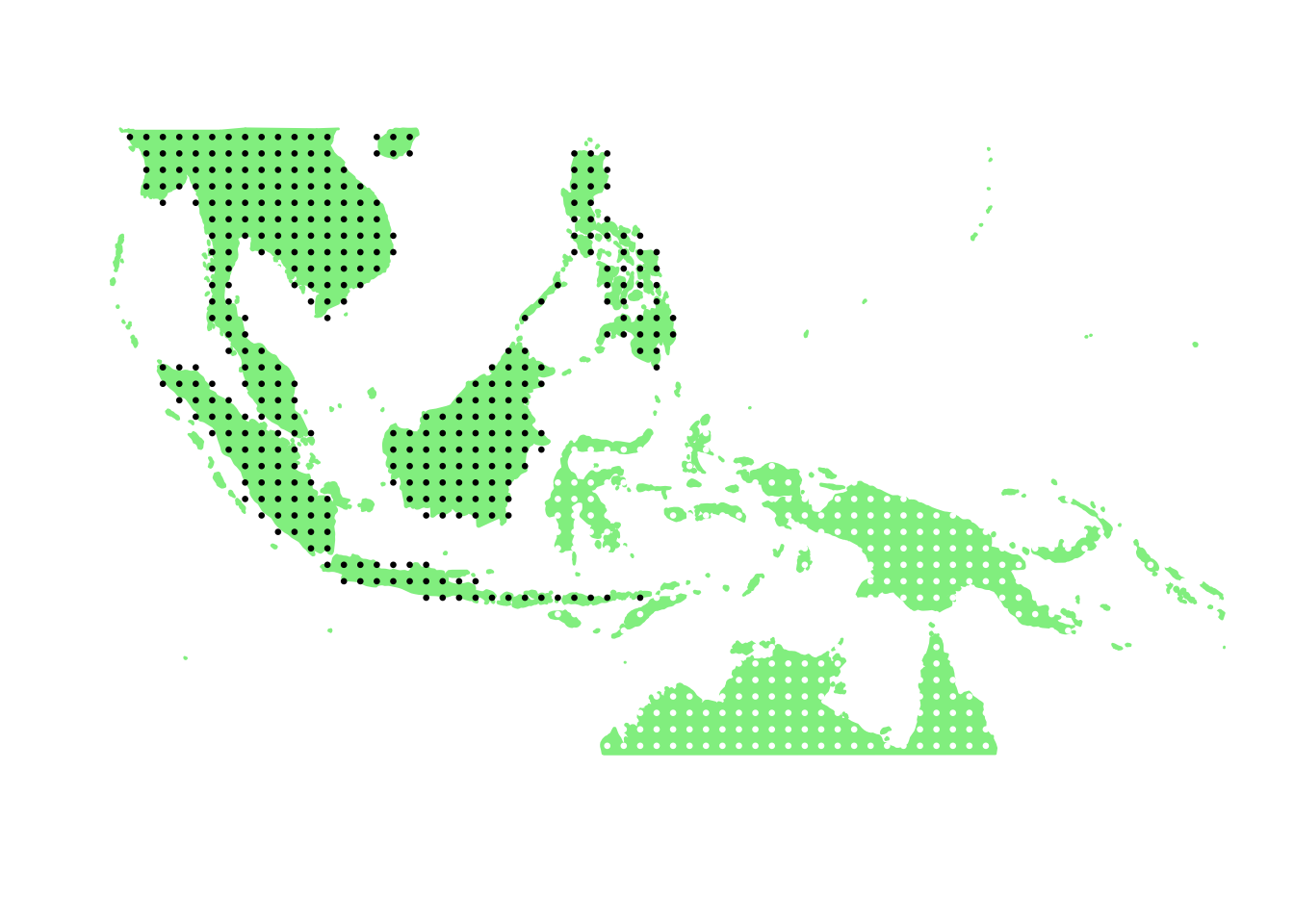
PlotAssemblageIdx(order(second_topic_scores, decreasing = TRUE)[7])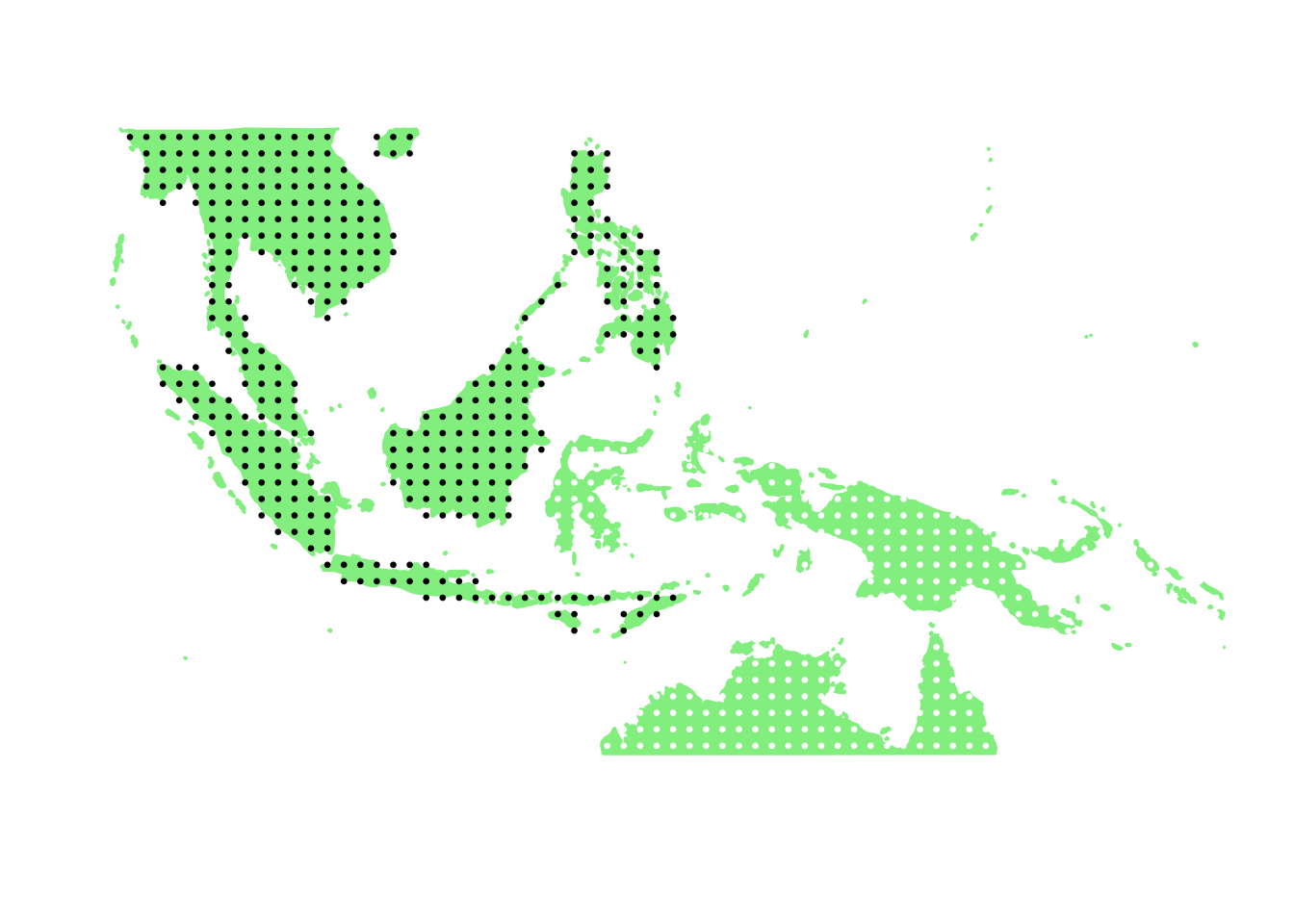
PlotAssemblageIdx(order(second_topic_scores, decreasing = TRUE)[8])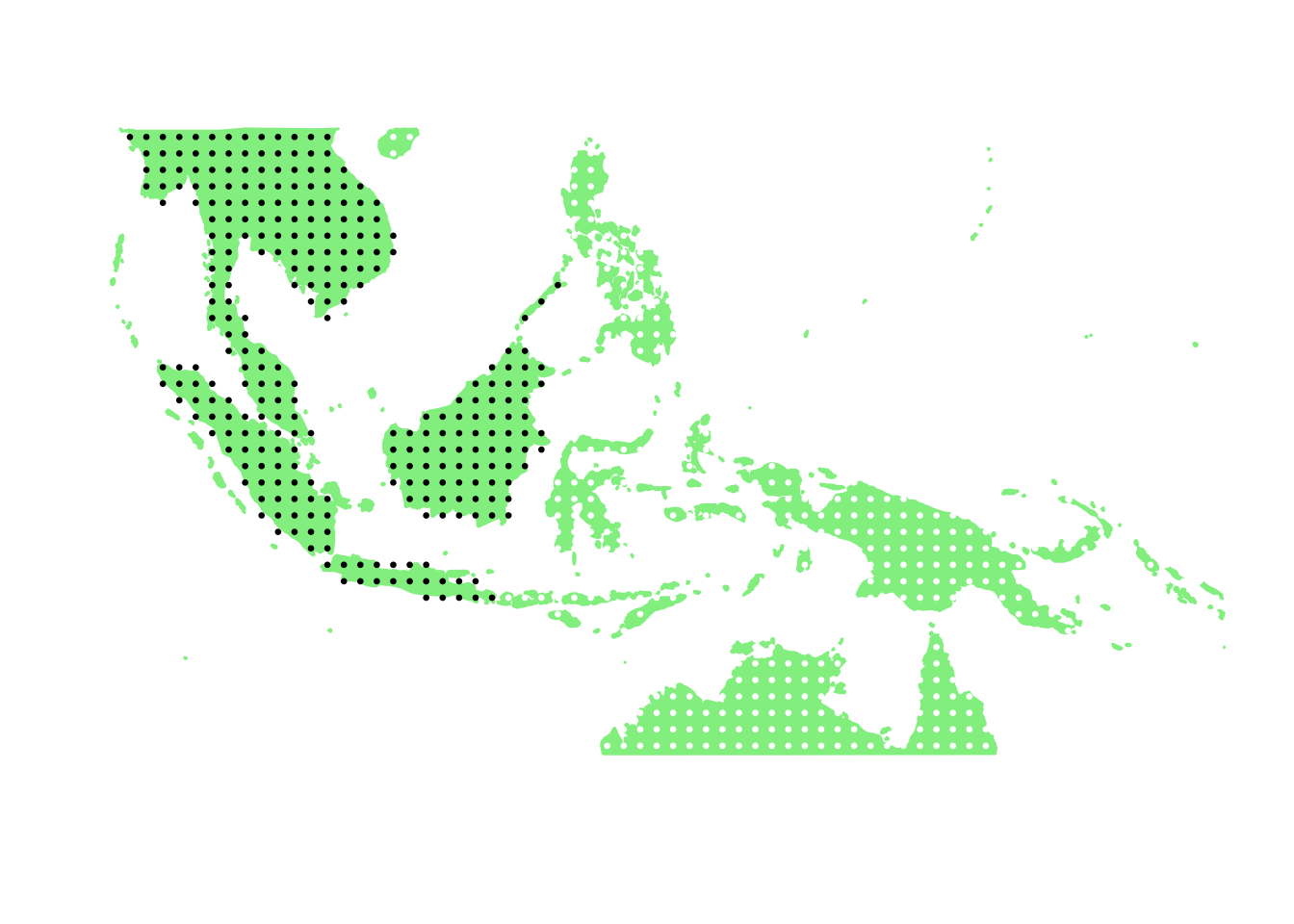
PlotAssemblageIdx(order(second_topic_scores, decreasing = TRUE)[9])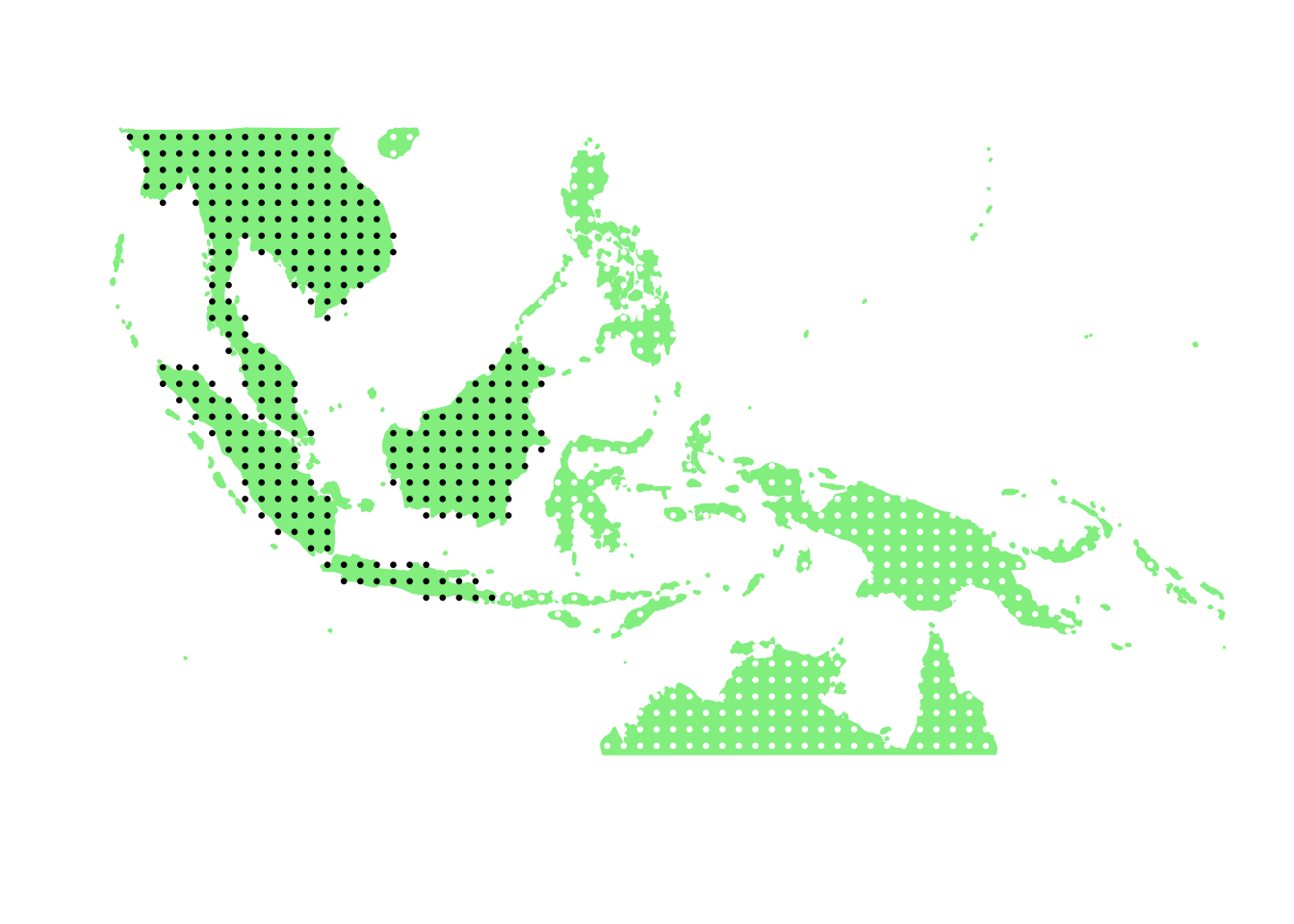
PlotAssemblageIdx(order(second_topic_scores, decreasing = TRUE)[10])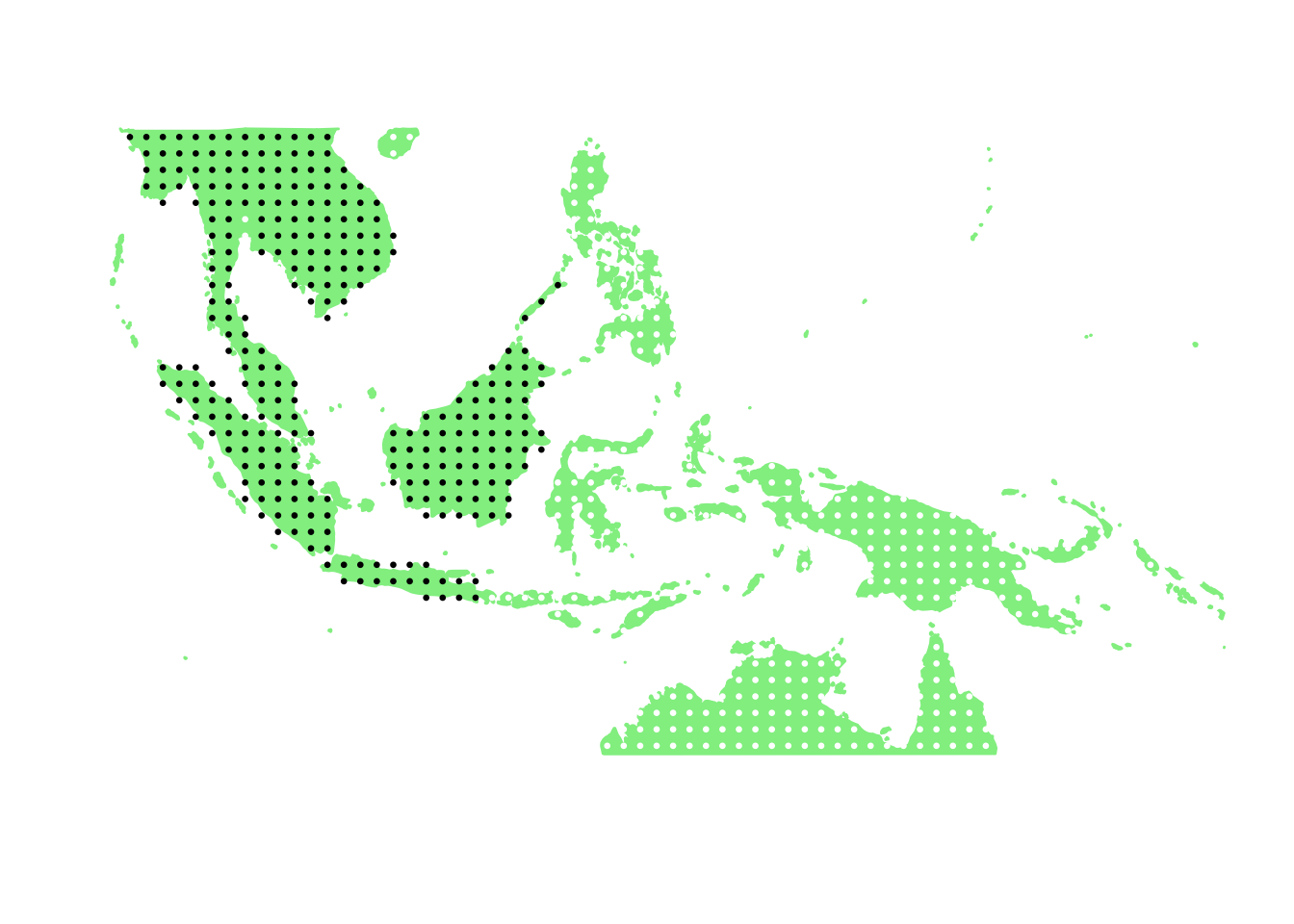
par(mfrow = c(5,2))
PlotAssemblageIdx(order(first_topic_scores, decreasing = TRUE)[1])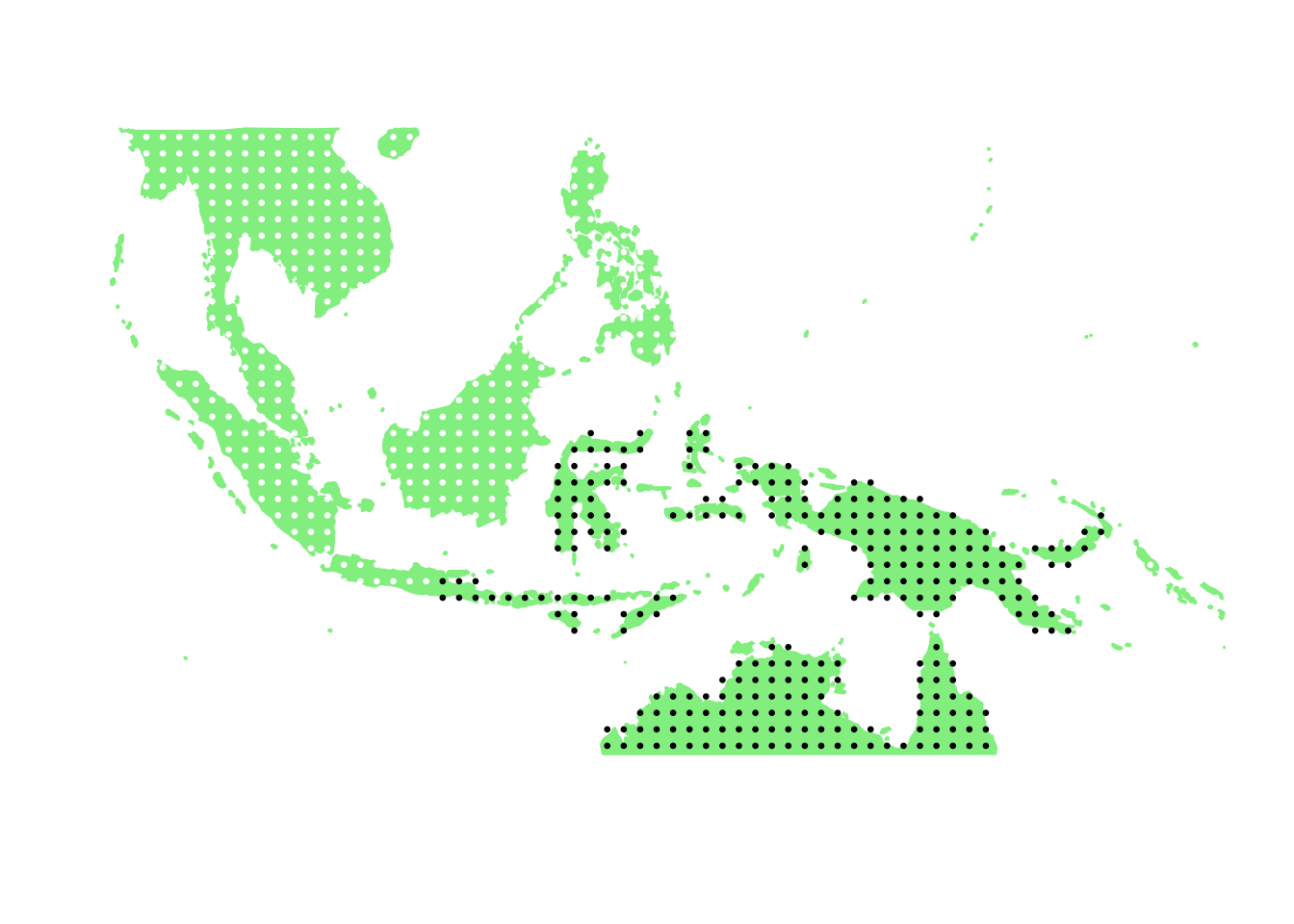
PlotAssemblageIdx(order(first_topic_scores, decreasing = TRUE)[2])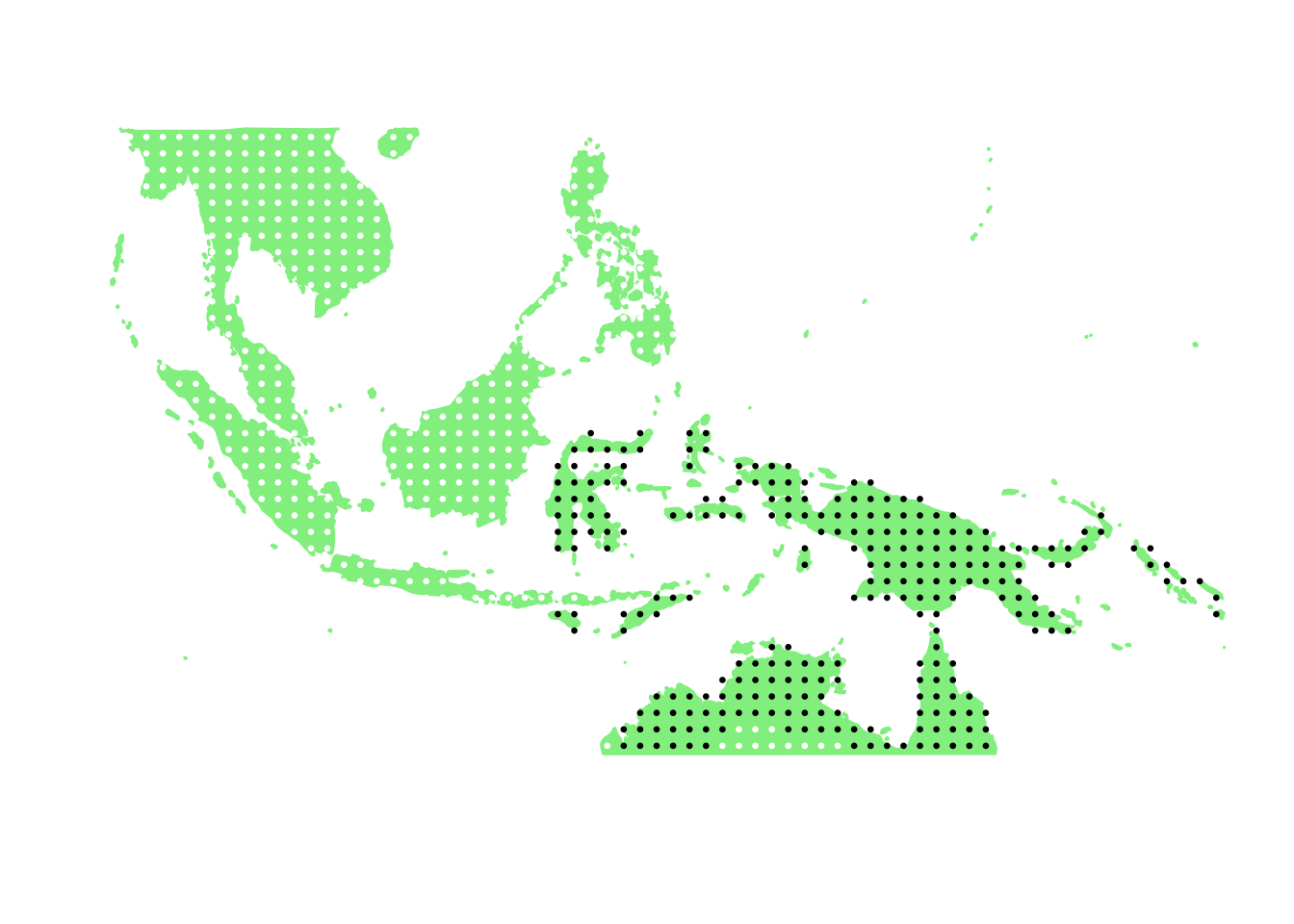
PlotAssemblageIdx(order(first_topic_scores, decreasing = TRUE)[3])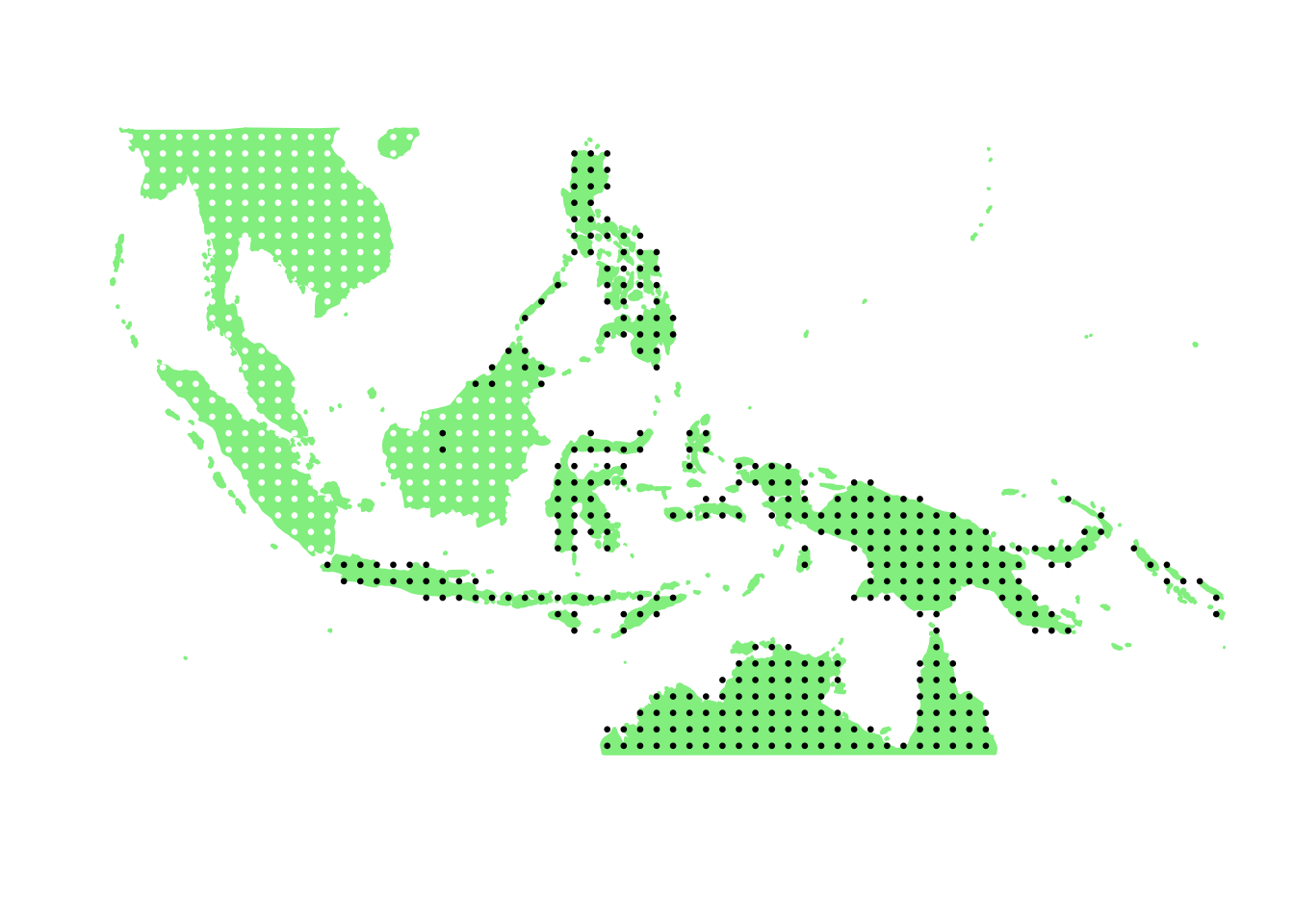
PlotAssemblageIdx(order(first_topic_scores, decreasing = TRUE)[4])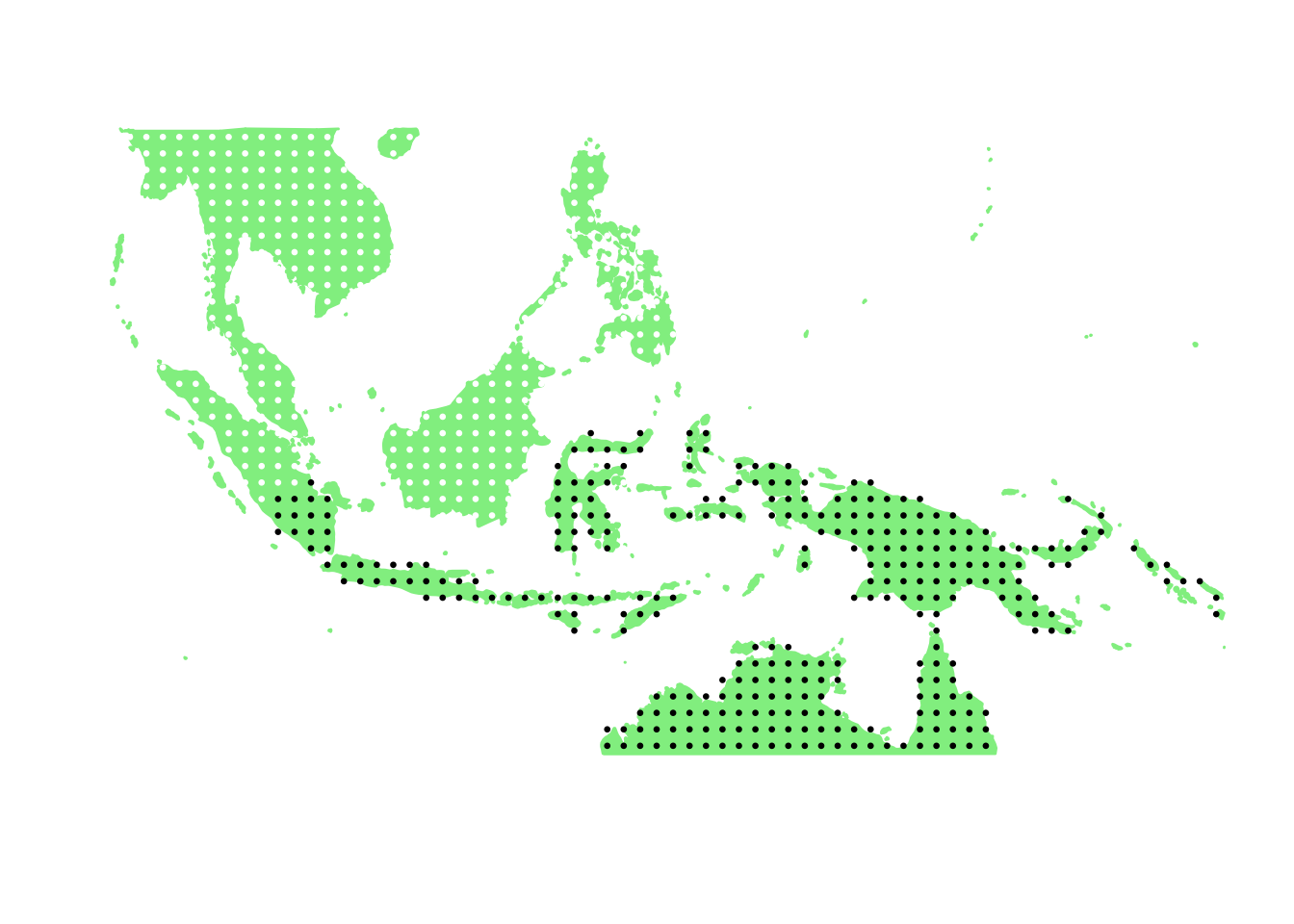
PlotAssemblageIdx(order(first_topic_scores, decreasing = TRUE)[5])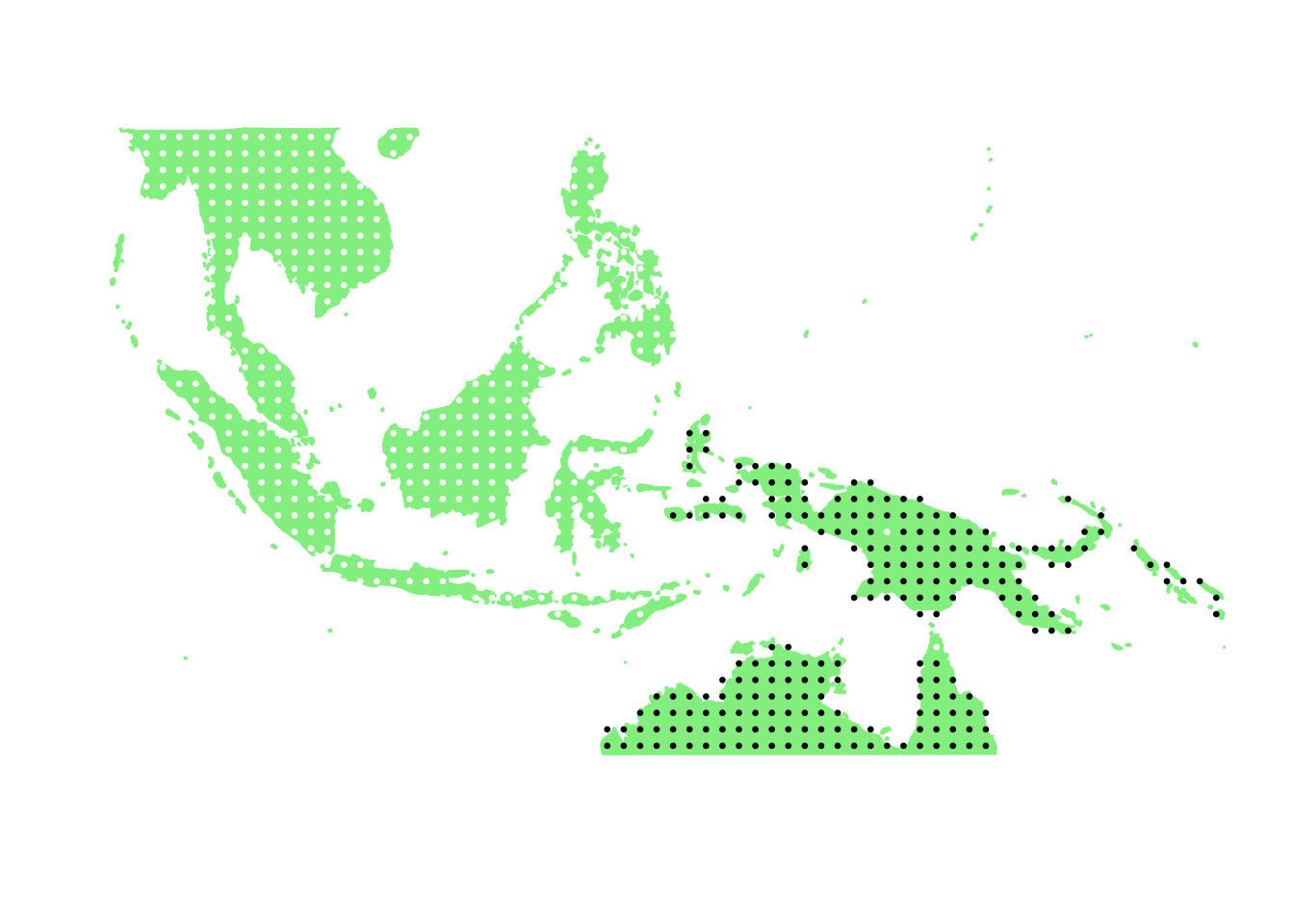
PlotAssemblageIdx(order(first_topic_scores, decreasing = TRUE)[6])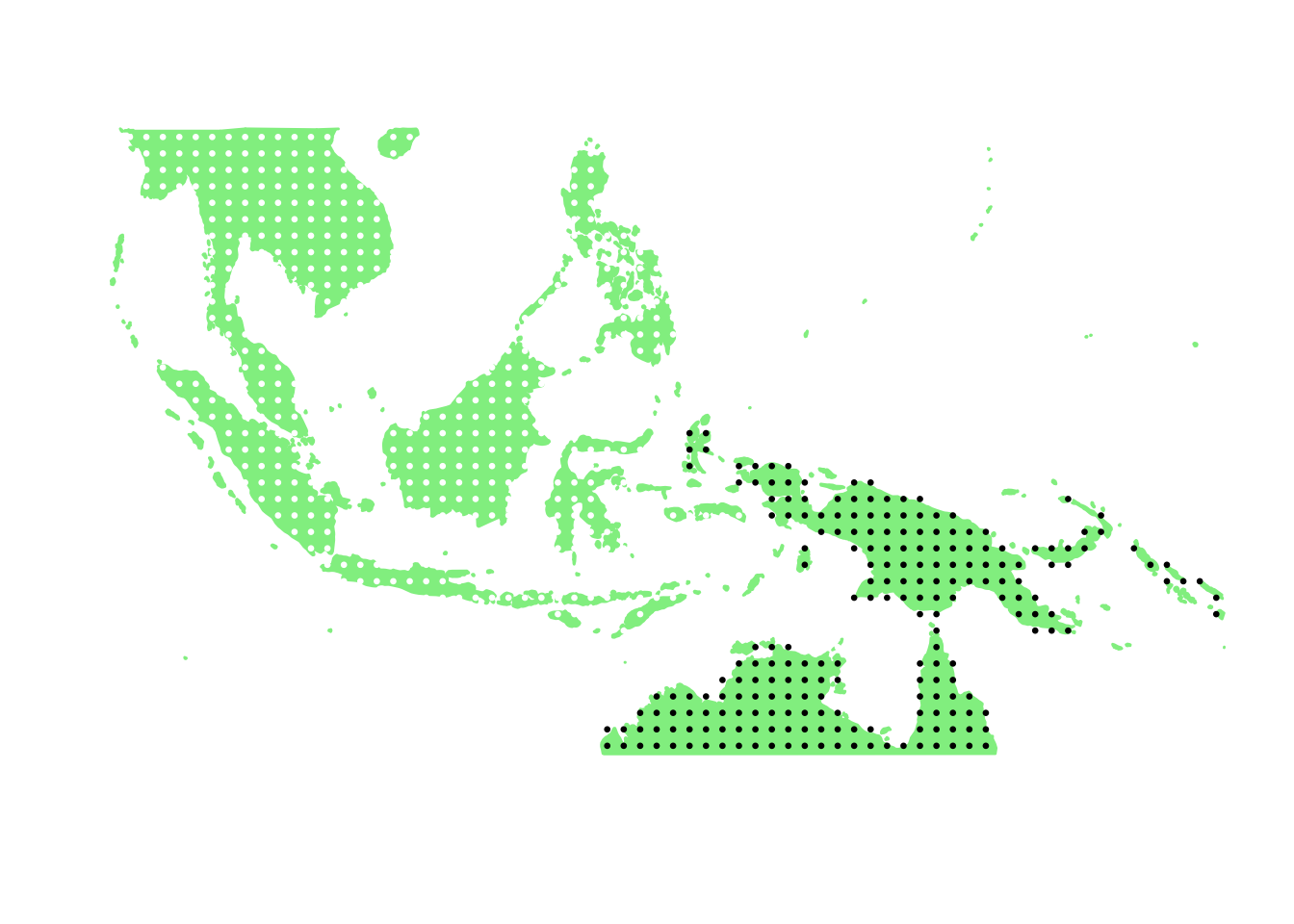
PlotAssemblageIdx(order(first_topic_scores, decreasing = TRUE)[7])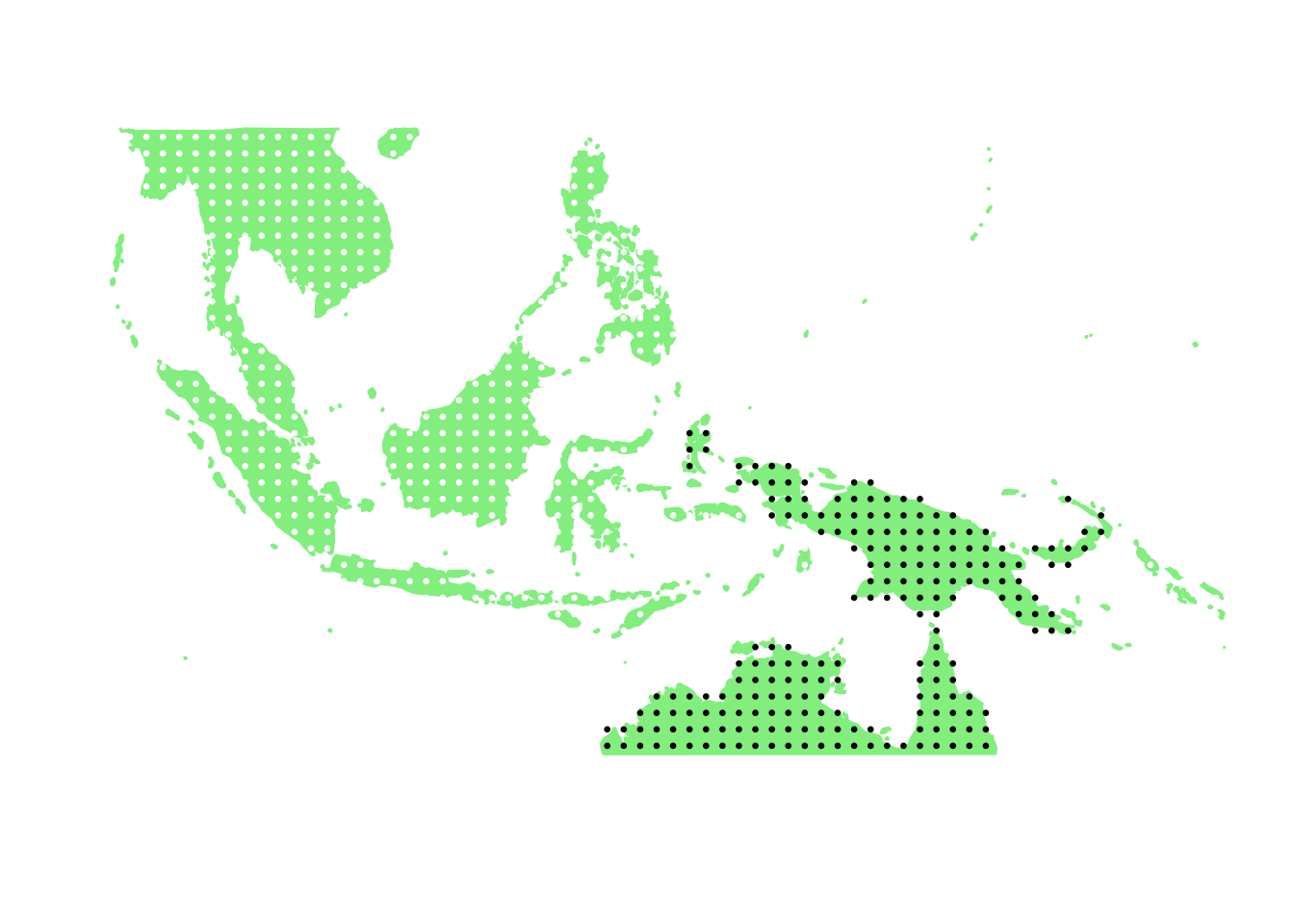
PlotAssemblageIdx(order(first_topic_scores, decreasing = TRUE)[8])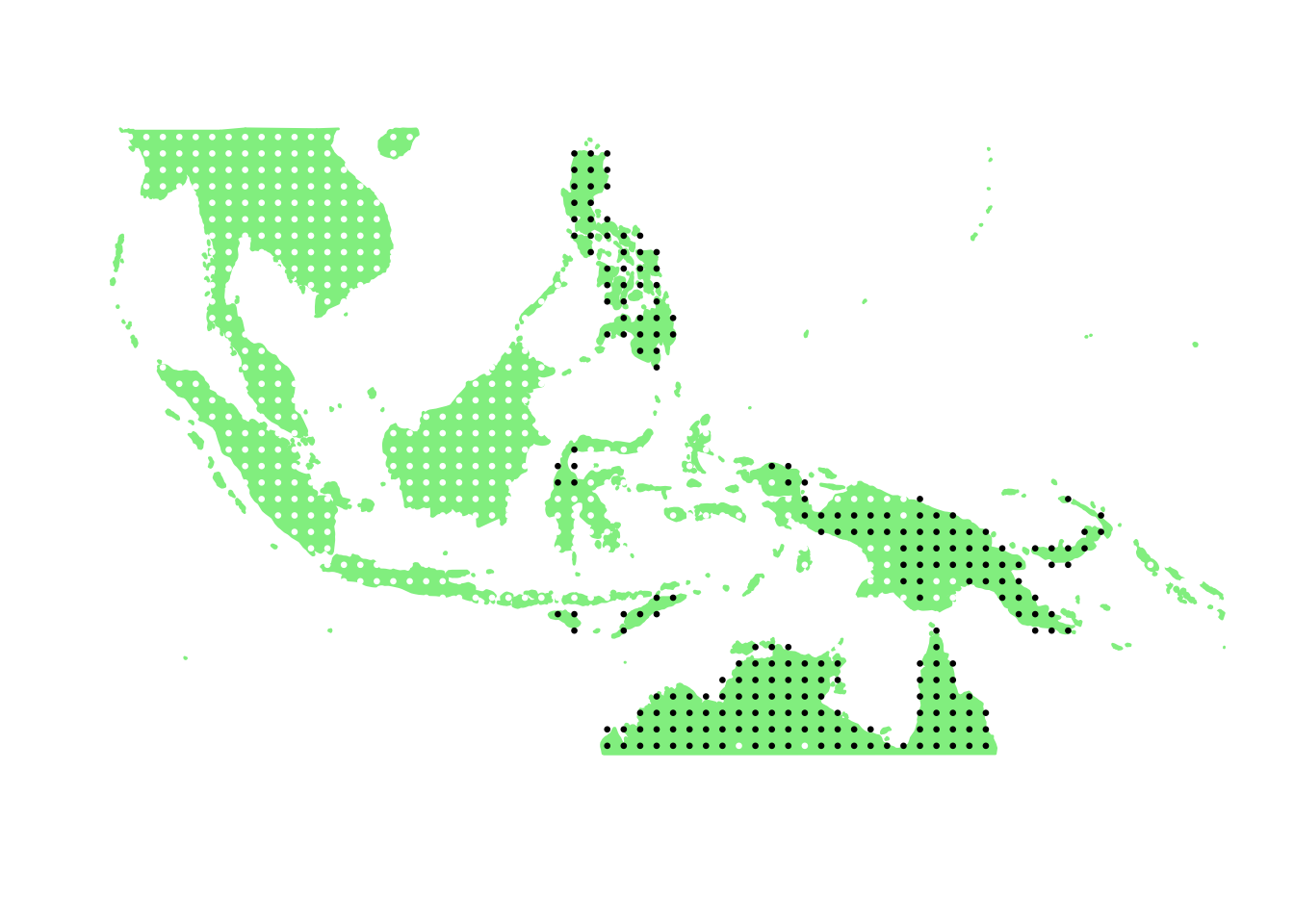
PlotAssemblageIdx(order(first_topic_scores, decreasing = TRUE)[9])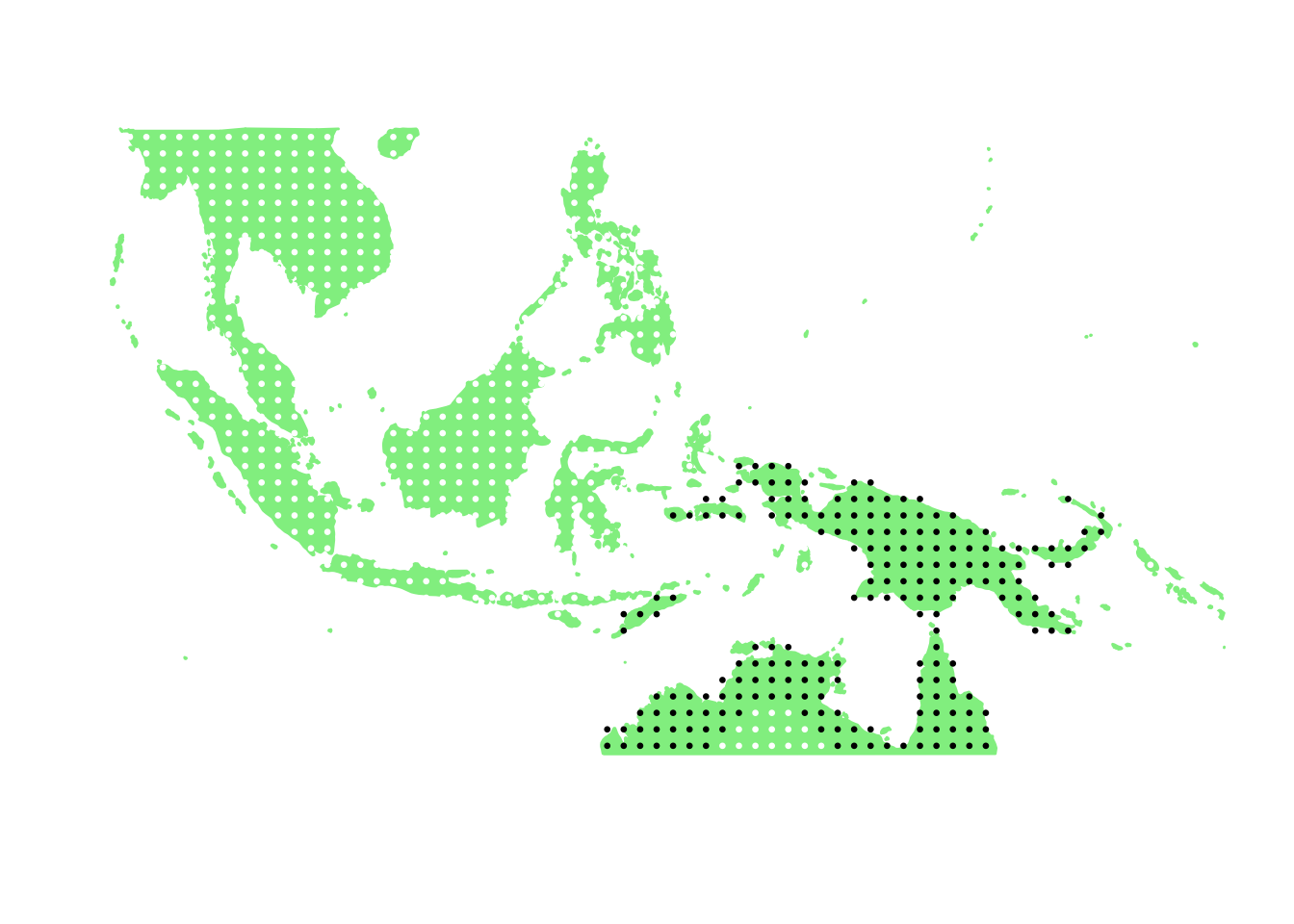
PlotAssemblageIdx(order(first_topic_scores, decreasing = TRUE)[10])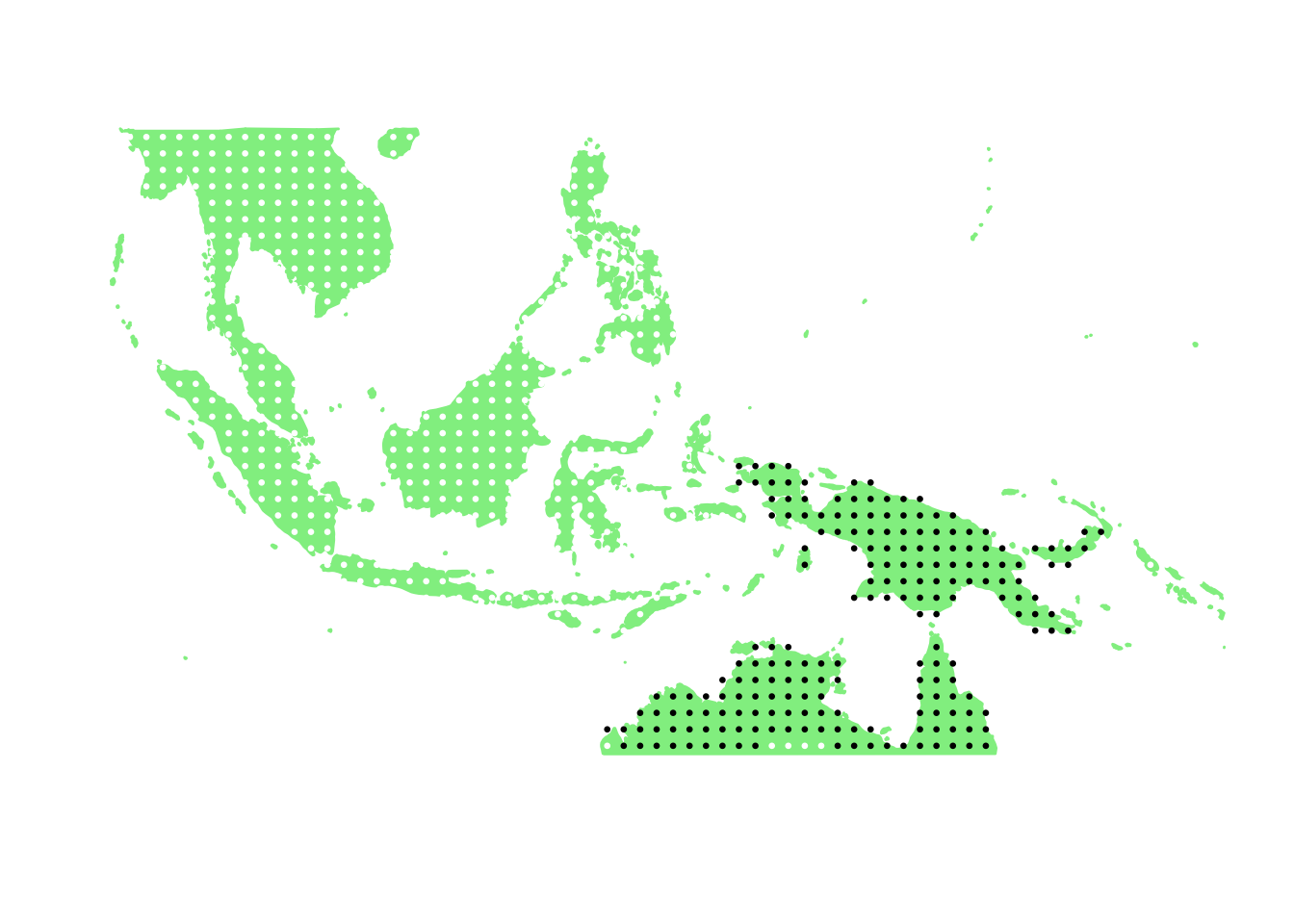
This R Markdown site was created with workflowr