Birds by cells versus cells by birds analysis
Kushal K Dey
12/16/2017
We compare the methClust model fit on the sites by bird species presence absence matrix with the bird species by sites presence absence matrix.
Plotting function of clusters
latlong <- get(load("../data/LatLongCells_frame.rda"))world_map <- map_data("world")
world_map <- world_map[world_map$region != "Antarctica",] # intercourse antarctica
p <- ggplot() + coord_fixed() +
xlab("") + ylab("")
#Add map to base plot
base_world_messy <- p + geom_map(data=world_map, map = world_map, aes(group=group, map_id=region), colour="white", fill="white", size=0.05, alpha=1/4)
cleanup <-
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
panel.background = element_rect(fill = 'white', colour = 'white'),
axis.line = element_line(colour = "white"), legend.position="none",
axis.ticks=element_blank(), axis.text.x=element_blank(),
axis.text.y=element_blank())
base_world <- base_world_messy + cleanupPlotAssemblageIdx <- function(dat){
dat <- cbind.data.frame(latlong, dat)
colnames(dat) <- c("Latitude", "Longitude", "Value")
map_data_coloured <-
base_world +
geom_point(data=dat,
aes(x=Latitude, y=Longitude, colour=Value), size=0.5) +
scale_colour_gradient(low = "white", high = "black")
map_data_coloured
}K = 2
(cells -> birds)
K2_global_topics <- get(load(file="../output/birds_meth_cluster_2.rda"))
topics_omega <- K2_global_topics$omega
PlotAssemblageIdx(topics_omega[,1])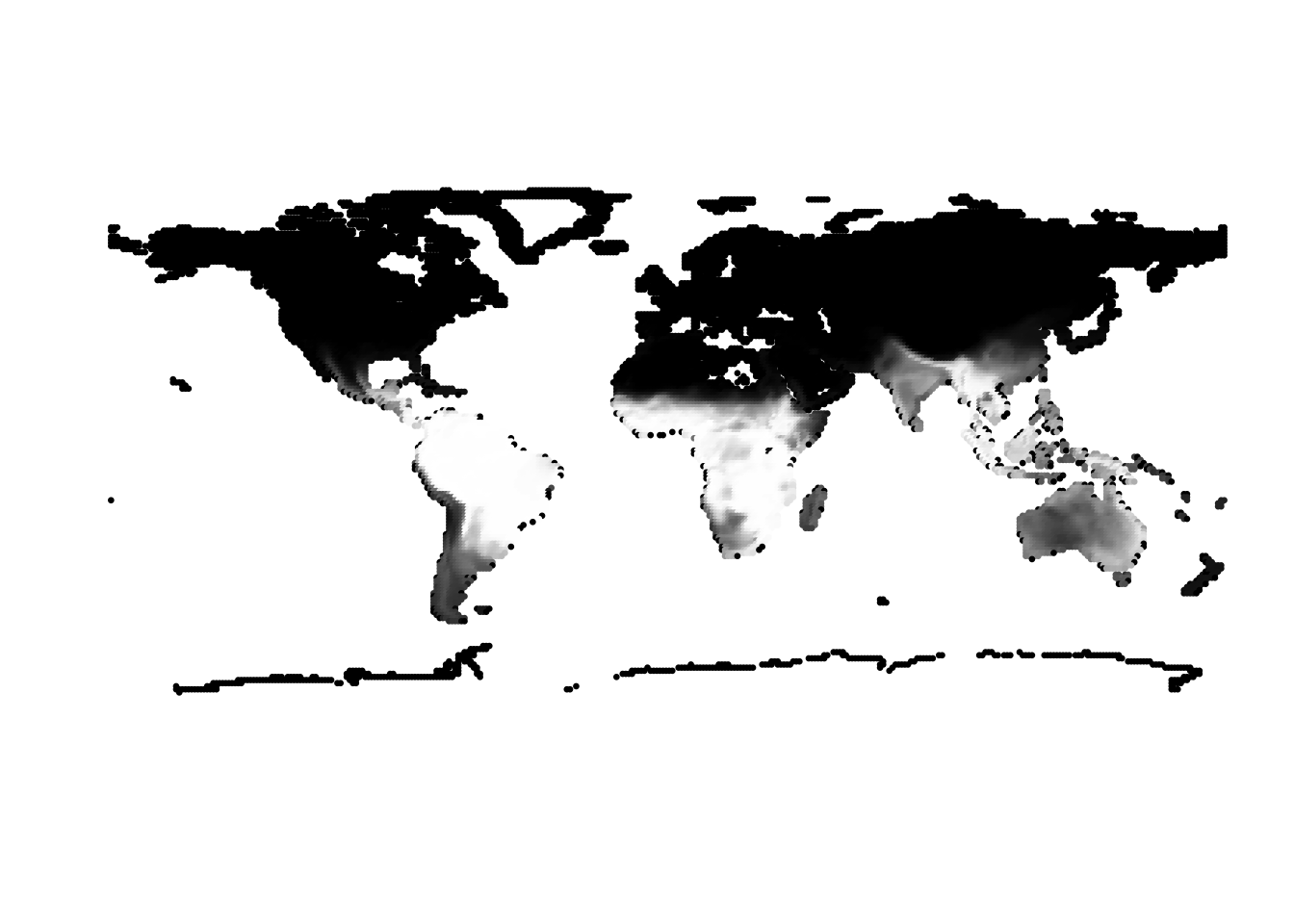
PlotAssemblageIdx(topics_omega[,2])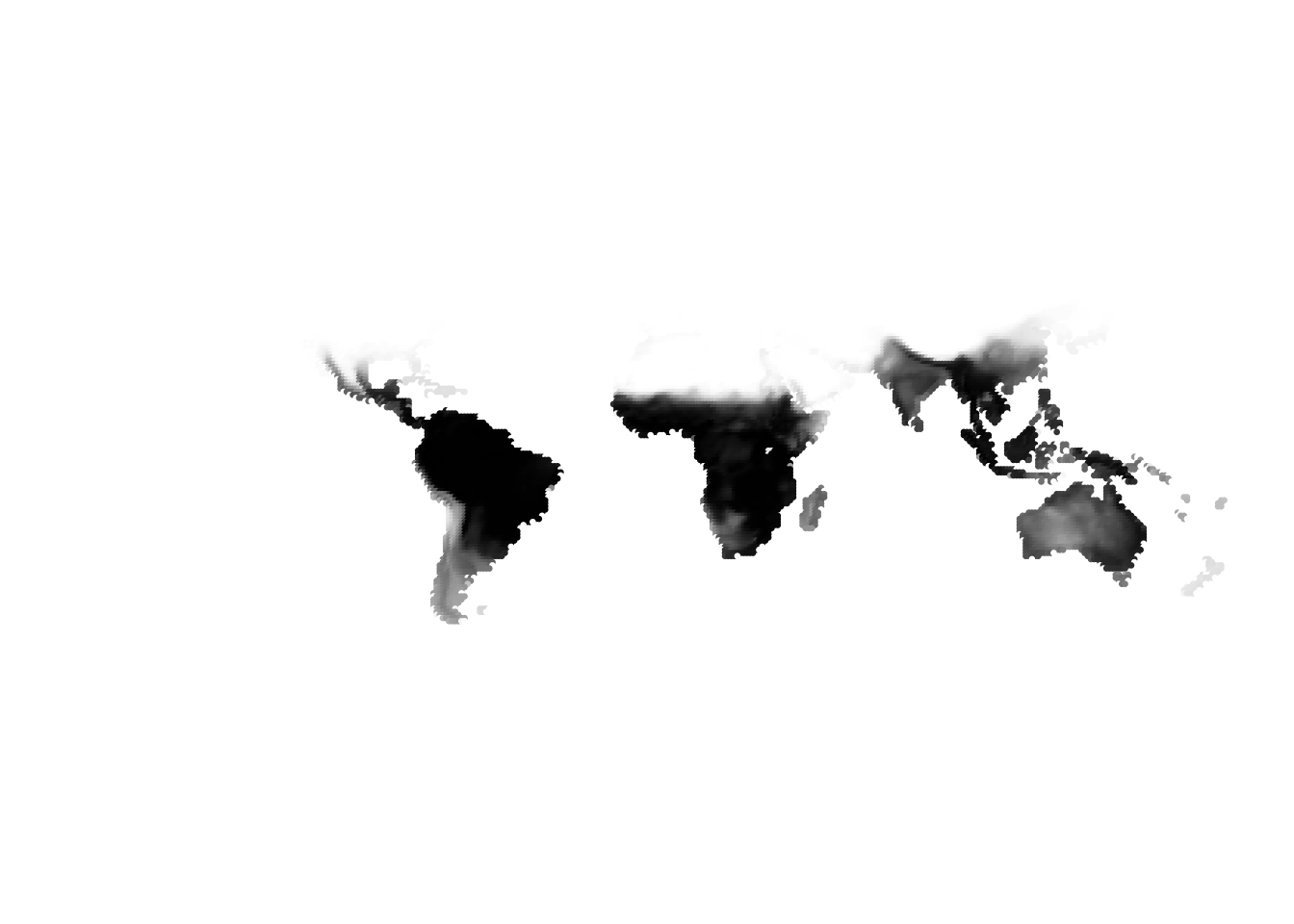
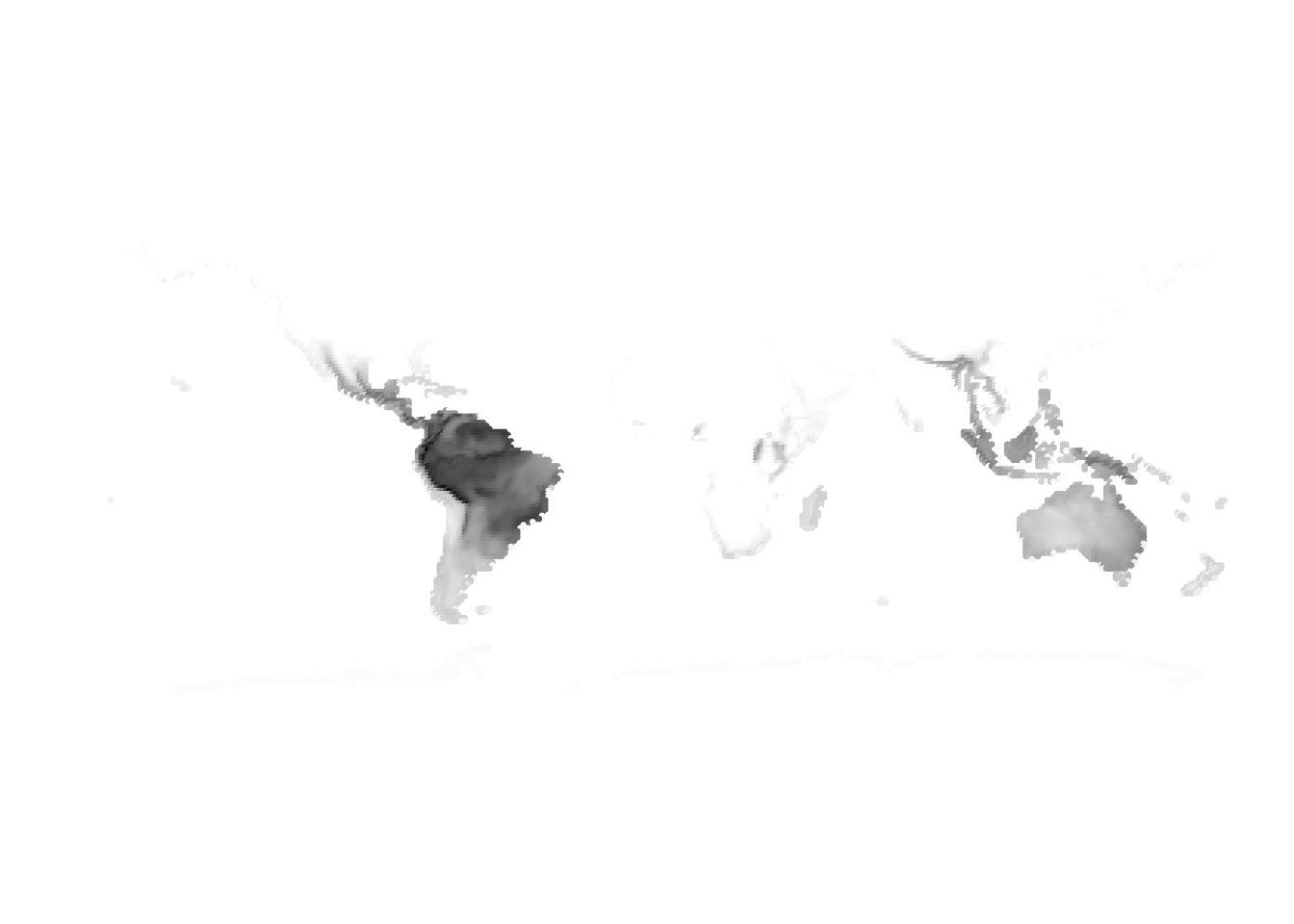
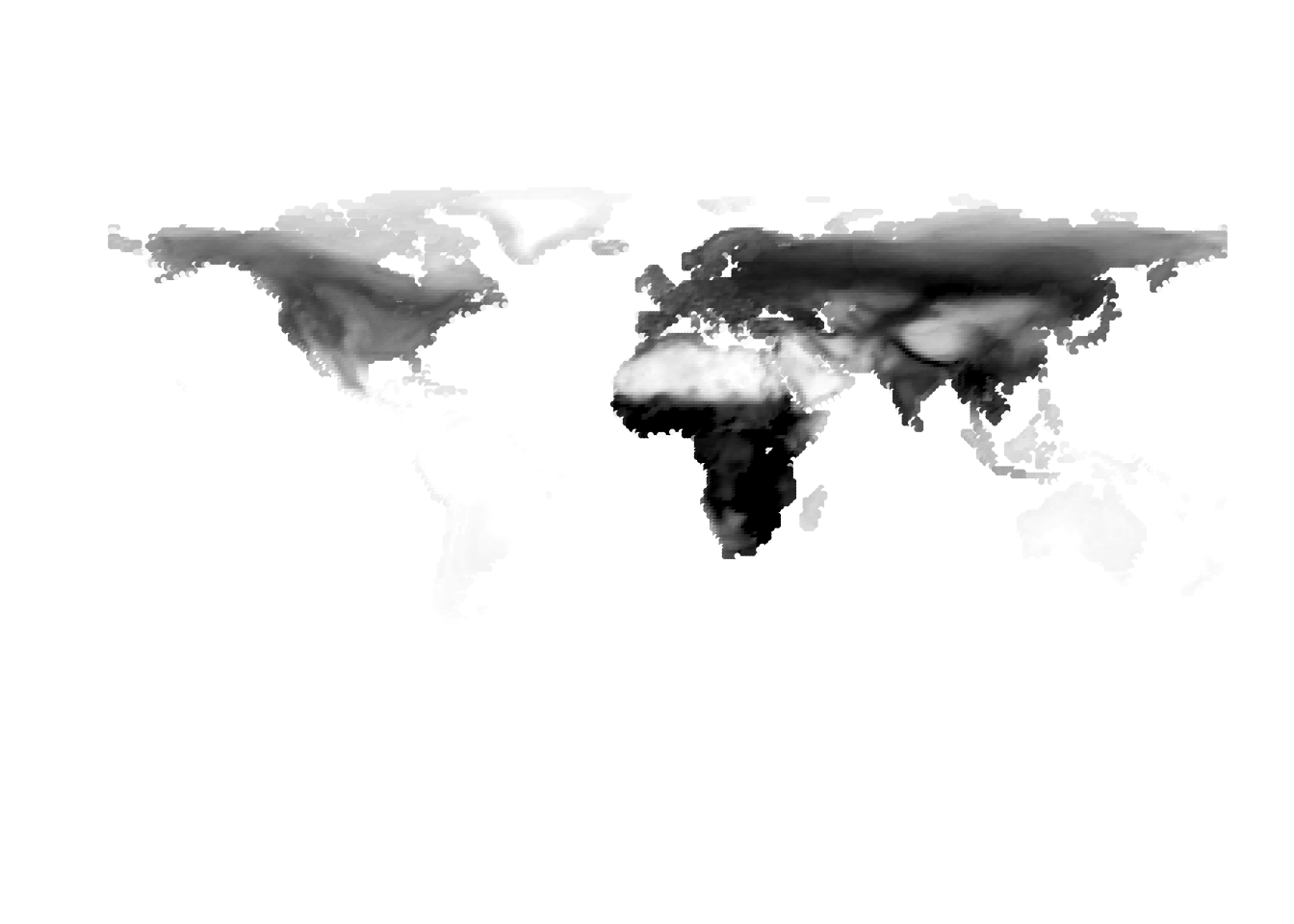
(birds -> cells)
Results for K = 2:
K = 3
(cells -> birds)
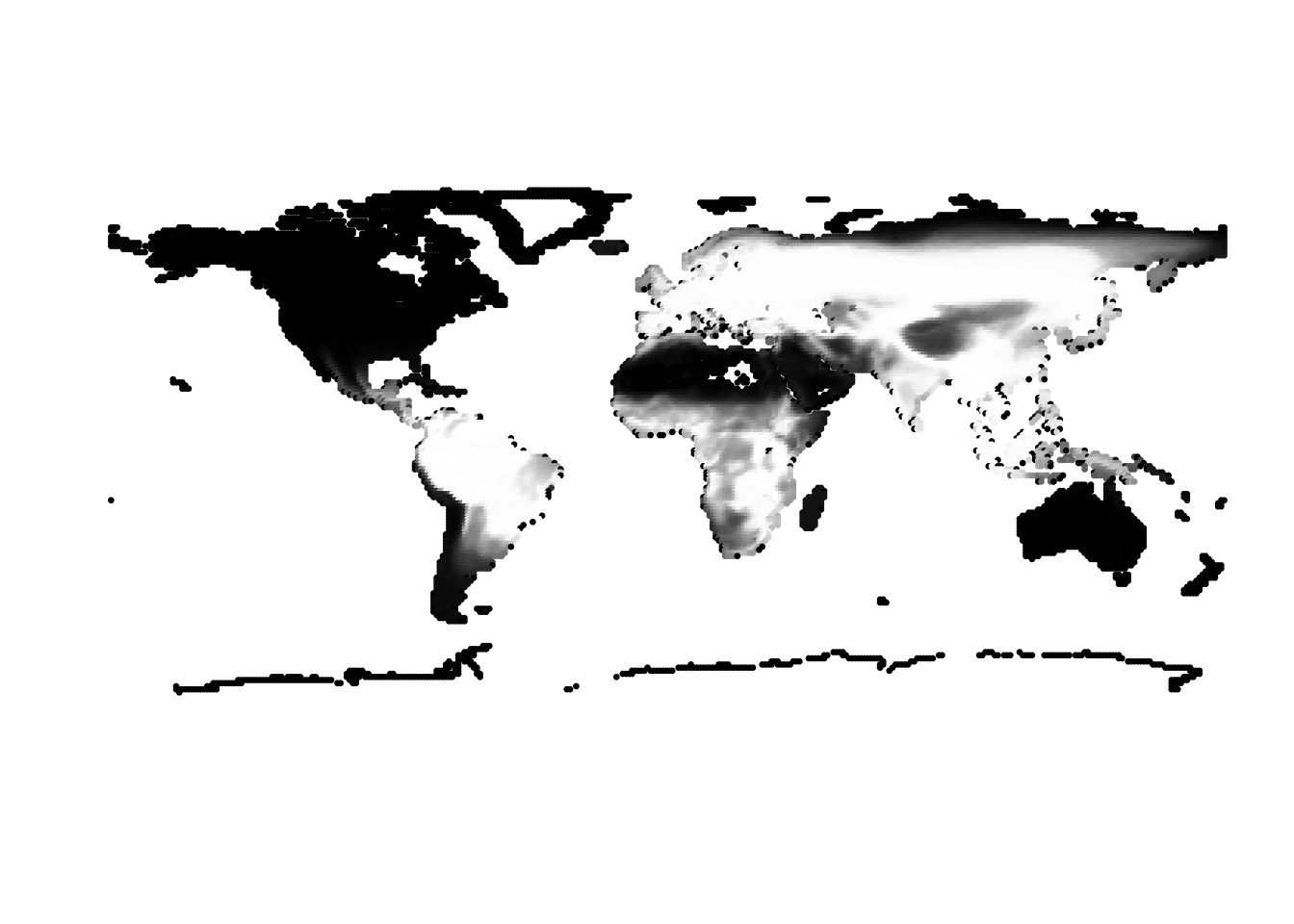


(birds -> cells)
Results for K = 3:

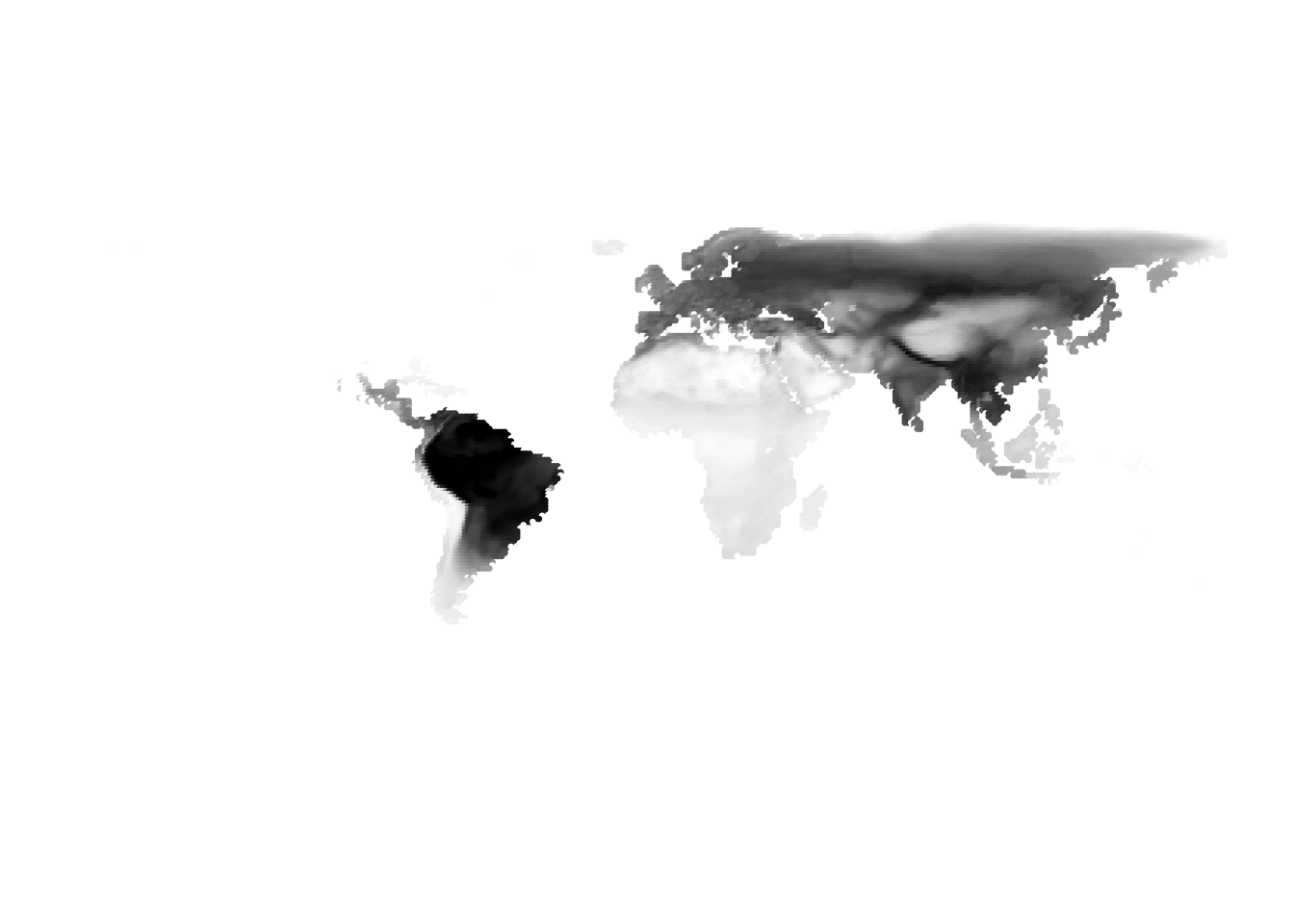
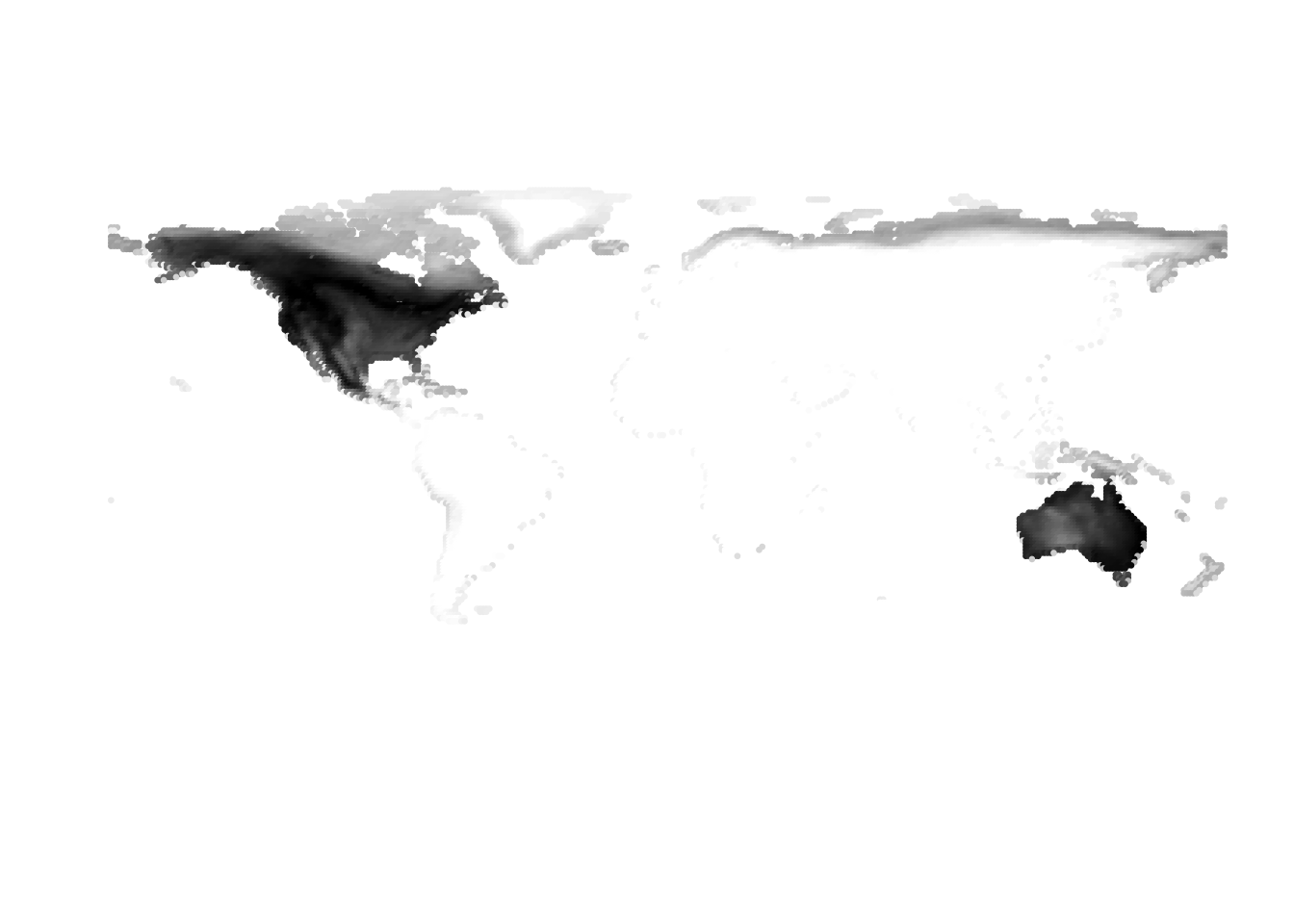
K = 5
(cells -> birds)
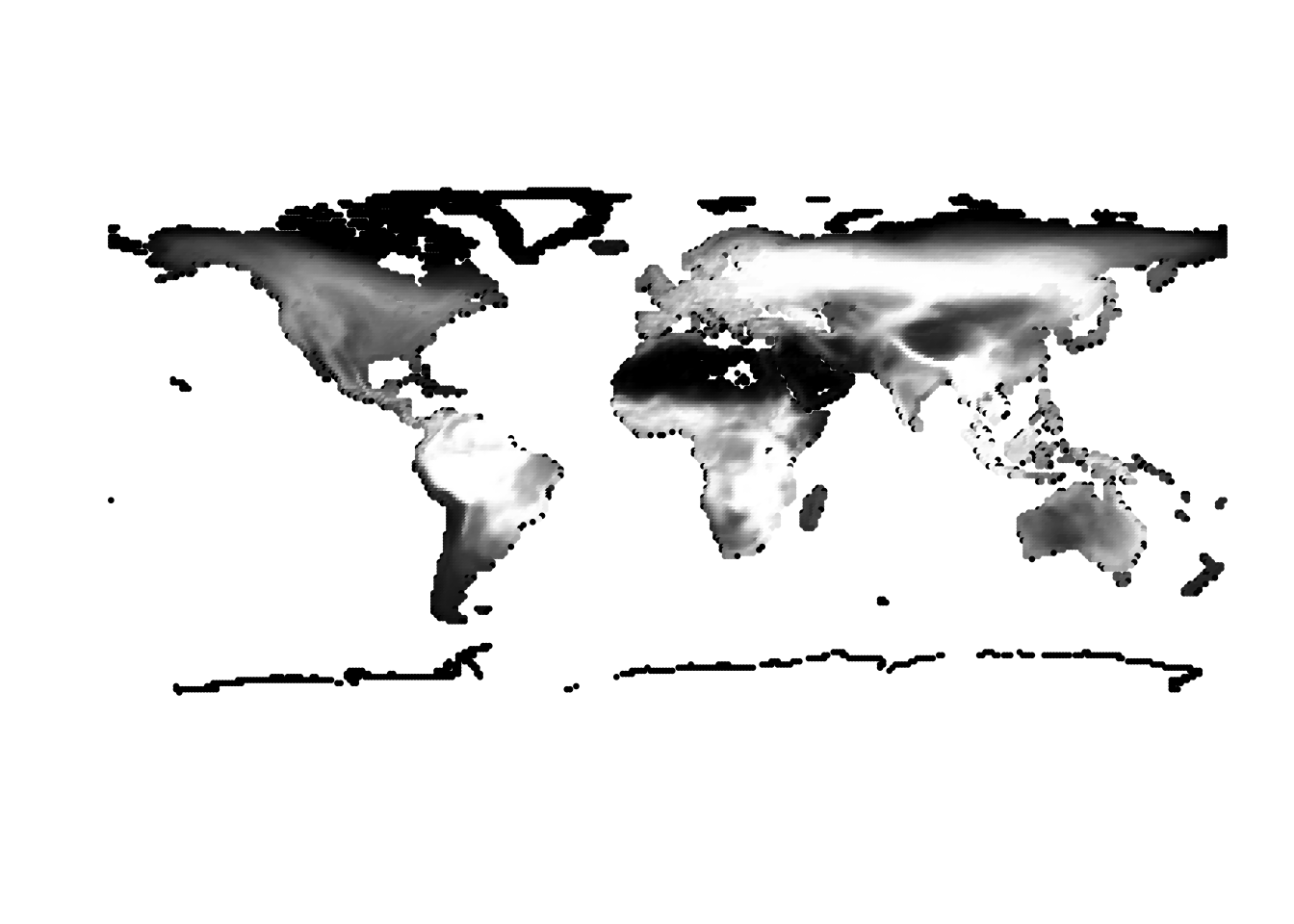




(birds -> cells)
Results for K = 5:





K = 10
(cells -> birds)
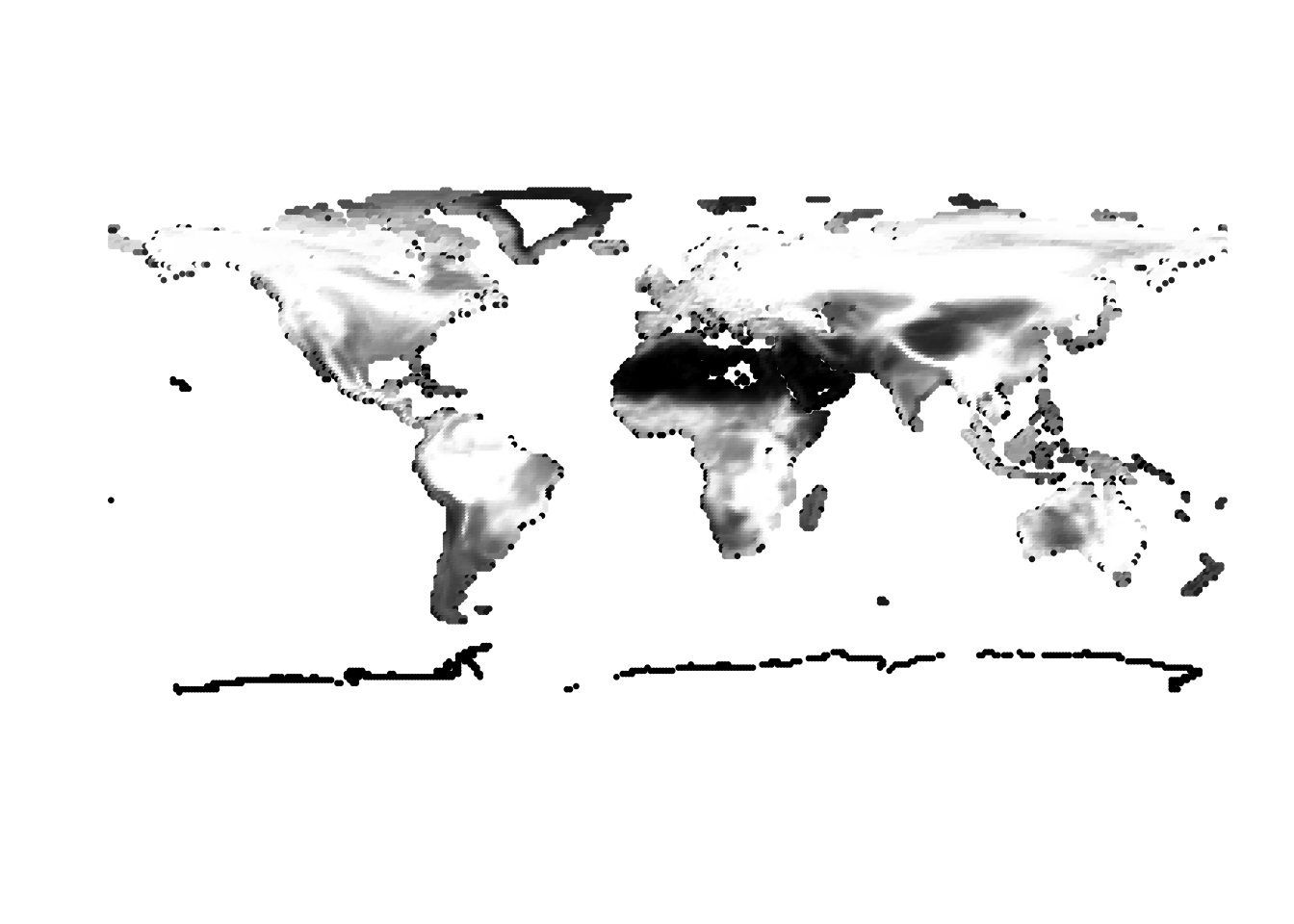









(birds -> cells)
Results for K = 10:







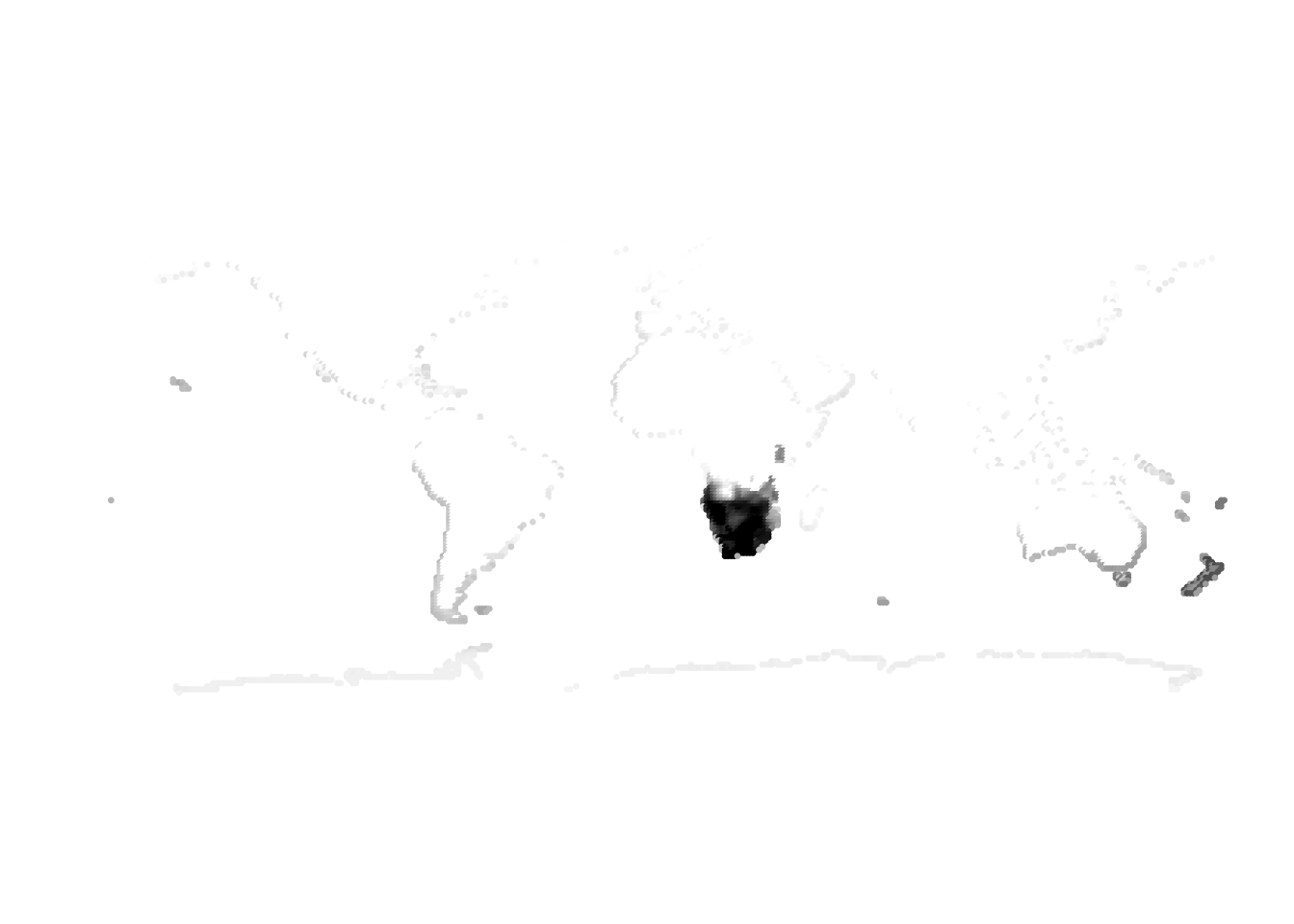


K = 11
(cells -> birds)
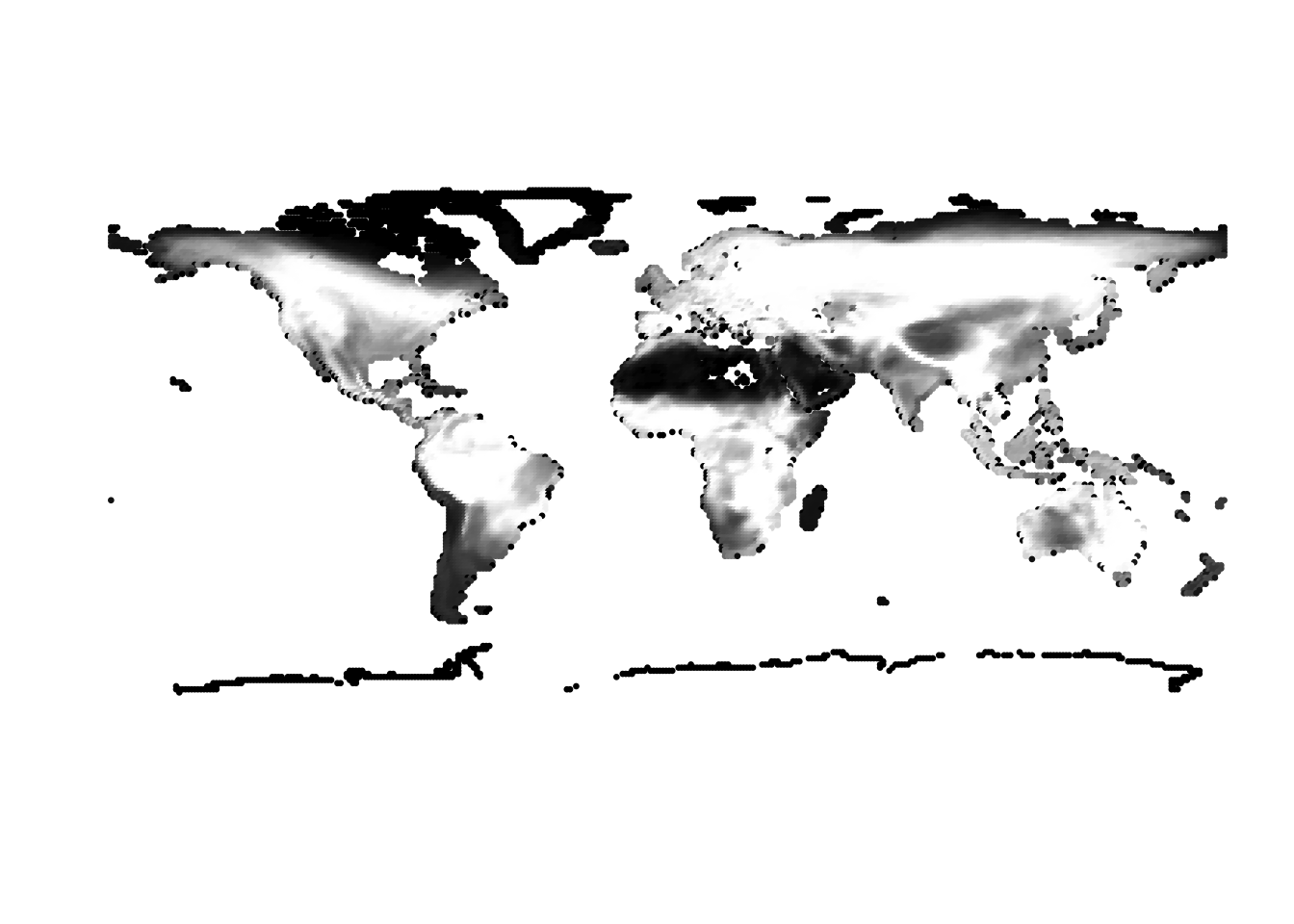










(birds -> cells)











This R Markdown site was created with workflowr