Supplementary Examples different structures
Kushal K. Dey and Rahul Mazumder
11/28/2019
Last updated: 2020-03-08
workflowr checks: (Click a bullet for more information)-
✖ R Markdown file: uncommitted changes
The R Markdown is untracked by Git. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can runwflow_publishto commit the R Markdown file and build the HTML. -
✔ Environment: empty
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
-
✔ Seed:
set.seed(20190721)The command
set.seed(20190721)was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible. -
✔ Session information: recorded
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
-
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.✔ Repository version: 47185fd
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can usewflow_publishorwflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.Ignored files: Ignored: .DS_Store Ignored: .Rhistory Ignored: .Rproj.user/ Ignored: docs/.DS_Store Ignored: draft/ Ignored: output/output_sparse/ Untracked files: Untracked: .gitignore~ Untracked: analysis/band_prec_robocov.Rmd Untracked: analysis/blood_corspan_robospan_probospan.Rmd Untracked: analysis/corspan_robospan_probospan.Rmd Untracked: analysis/diff_robospan_examples.Rmd Untracked: analysis/eigenvalues_hub.Rmd Untracked: analysis/figure2.Rmd Untracked: analysis/gtex_analysis_robocov_examples.Rmd Untracked: analysis/gtex_pRobocov_pRobospan.Rmd Untracked: analysis/gtex_predictive_robocov.Rmd Untracked: analysis/gtex_rank_imputation.Rmd Untracked: analysis/gtex_robocov.Rmd Untracked: analysis/gtex_robocov_ex_robospan.Rmd Untracked: analysis/housekeeping_PPI_MR_enrichment.Rmd Untracked: analysis/hub_robocov.Rmd Untracked: analysis/mean_robocov.Rmd Untracked: analysis/pLI_shet_robospan_probospan.Rmd Untracked: analysis/predictive_analytics_gtex.Rmd Untracked: analysis/robocov_demo.Rmd Untracked: analysis/robospan_blood_compare.Rmd Untracked: analysis/robospan_corr.Rmd Untracked: analysis/summary_correlation_structure.Rmd Untracked: analysis/supp_figure_simulation.Rmd Untracked: analysis/toeplitz_robocov.Rmd Untracked: code/ robocov_sim.R Untracked: code/FDR_Robospan.R Untracked: code/Joints_Nov24.R Untracked: code/Untitled.R Untracked: code/annot_size.R Untracked: code/band_prec_ocor_sim.R Untracked: code/band_prec_pcor_sim.R Untracked: code/band_prec_sim.R Untracked: code/band_prec_sim2.R Untracked: code/bandprec_cor_sim.R Untracked: code/baseline_strategies.R Untracked: code/corr_span.R Untracked: code/correlation_gene_scores.R Untracked: code/gene_score_all_strategies.R Untracked: code/gtex_robocov.R Untracked: code/gtex_robocov_precision.R Untracked: code/hub_prec_cor_sim.R Untracked: code/hub_sim.R Untracked: code/hub_sim2.R Untracked: code/hub_sim_sparse.R Untracked: code/joint_robocov_model.R Untracked: code/joints_robocov.R Untracked: code/many_tau_star.R Untracked: code/maxHiCgene.R Untracked: code/meta_enrich.R Untracked: code/meta_single_enrich.R Untracked: code/postprocess_hub_toeplitz.R Untracked: code/predictive_analytics_hub.R Untracked: code/predictive_analytics_toeplitz.R Untracked: code/robocov_gtex.R Untracked: code/robocov_gtex_blood.R Untracked: code/robocov_gtex_brain.R Untracked: code/sim_results.R Untracked: code/single_tau_star.R Untracked: code/toeplitz_prec_cor_sim.R Untracked: code/toeplitz_sim.R Untracked: code/toeplitz_sim2.R Untracked: code/toeplitz_sim_sparse.R Untracked: data/Cor_pairwise_all_genes.rda Untracked: data/Gene_Scores/ Untracked: data/Robocov_Box_all_genes.rda Untracked: data/Robocov_Precision_all_genes.rda Untracked: data/gene_names_GTEX_V6.txt Untracked: data/housekeeping_genes.txt Untracked: data/person_tissue_genes_voom.rda Untracked: docs/figure/ Untracked: sim_code/ Unstaged changes: Modified: .gitignore Modified: analysis/index.Rmd
library(corrplot)corrplot 0.84 loadedrobocov_gtex = get(load("/Users/kushaldey/Documents/Robocov-pages/data/Robocov_Box_all_genes.rda"))
dim(robocov_gtex)[1] 53 53 16069robospan = apply(robocov_gtex, 3, sum)/(53*53)
plot(density(robospan), xlab = "Robospan score", ylab = "density")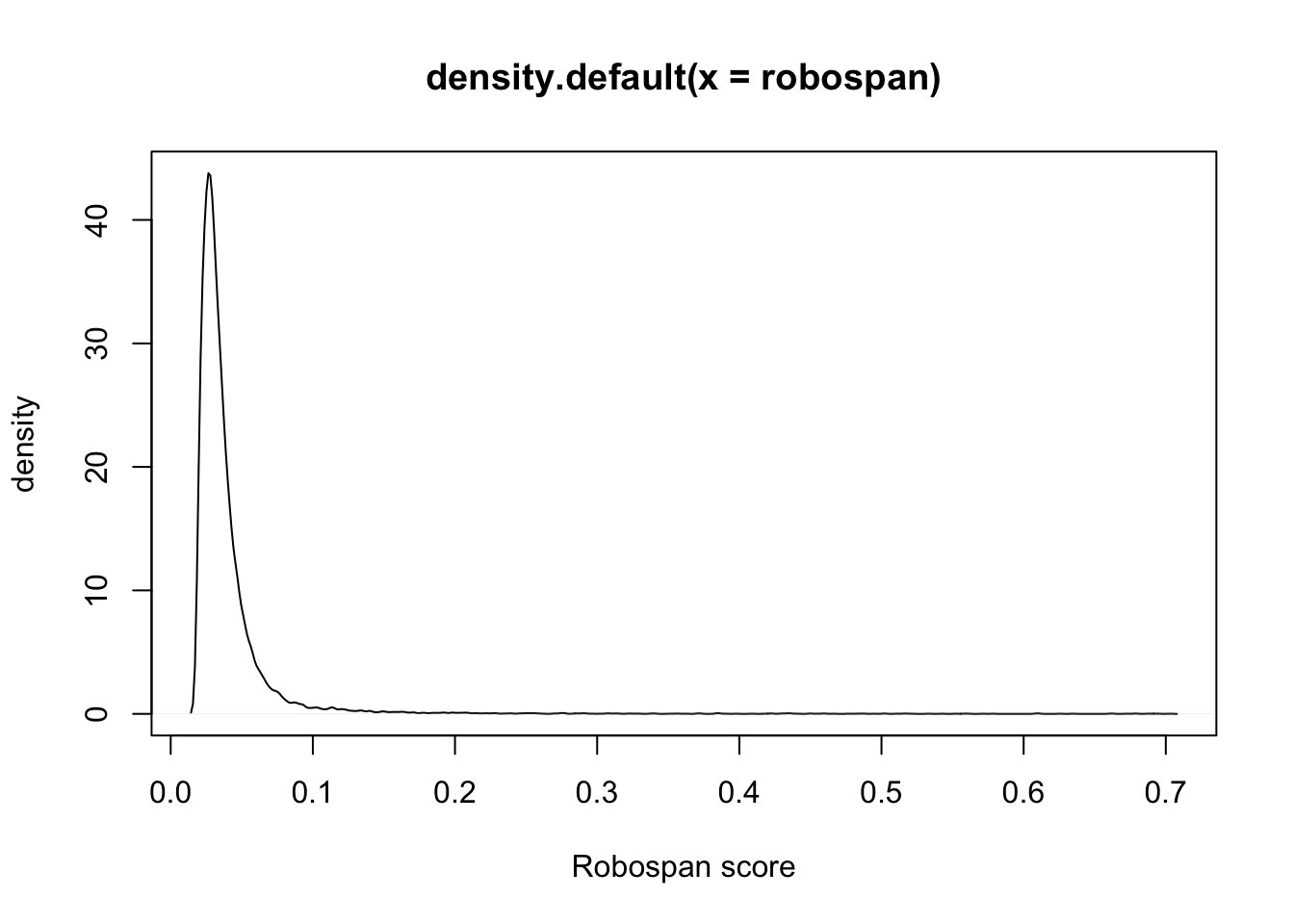
robospan[order(robospan, decreasing = T)[1:10]]ENSG00000134184 ENSG00000226278 ENSG00000184674 ENSG00000163682
0.7032426 0.6950521 0.6912778 0.6872412
ENSG00000233927 ENSG00000260246 ENSG00000204792 ENSG00000183298
0.6794906 0.6776832 0.6722359 0.6624388
ENSG00000196436 ENSG00000271581
0.6610709 0.6344899 robospan[order(robospan, decreasing = F)[1:20]]ENSG00000235795 ENSG00000257307 ENSG00000242100 ENSG00000228205
0.01889093 0.01890535 0.01891395 0.01892251
ENSG00000197830 ENSG00000227344 ENSG00000268543 ENSG00000258782
0.01892328 0.01892549 0.01895040 0.01895996
ENSG00000188505 ENSG00000180389 ENSG00000251791 ENSG00000266501
0.01896545 0.01896681 0.01897532 0.01898412
ENSG00000236992 ENSG00000132698 ENSG00000269038 ENSG00000224415
0.01899052 0.01900245 0.01900308 0.01900340
ENSG00000242571 ENSG00000213590 ENSG00000240254 ENSG00000180211
0.01900925 0.01901623 0.01901845 0.01902939 corrplot(robocov_gtex[,,"ENSG00000242571"], diag = TRUE,
col = colorRampPalette(c("lightblue4", "lightblue2", "white", "indianred1", "indianred3"))(200),
tl.pos = "ld", tl.cex = 0.8, tl.col = "black",
rect.col = "white",na.label.col = "white",
method = "color", type = "lower", tl.srt=45)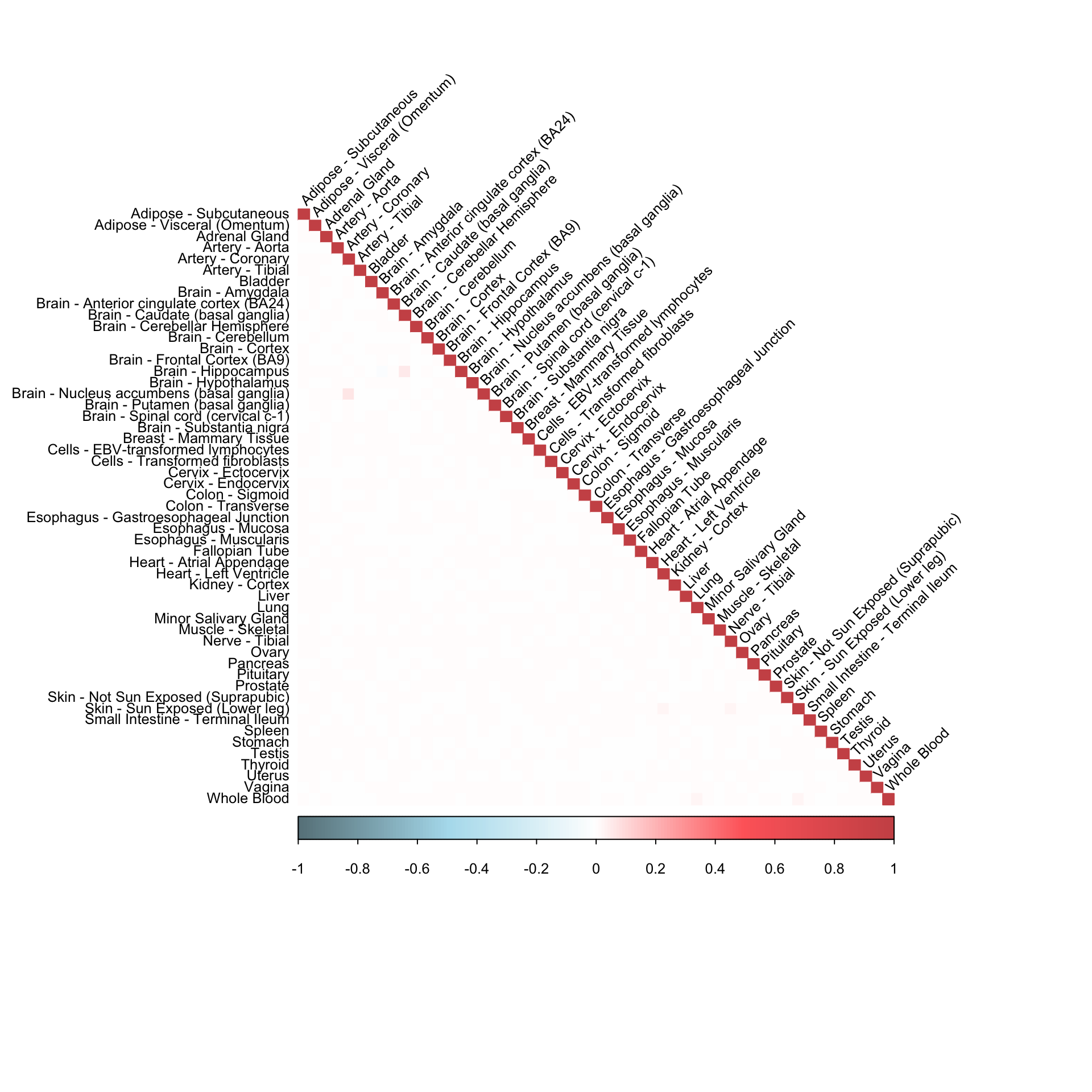
corrplot(robocov_gtex[,,"ENSG00000134184"], diag = TRUE,
col = colorRampPalette(c("lightblue4", "lightblue2", "white", "indianred1", "indianred3"))(200),
tl.pos = "ld", tl.cex = 0.8, tl.col = "black",
rect.col = "white",na.label.col = "white",
method = "color", type = "lower", tl.srt=45)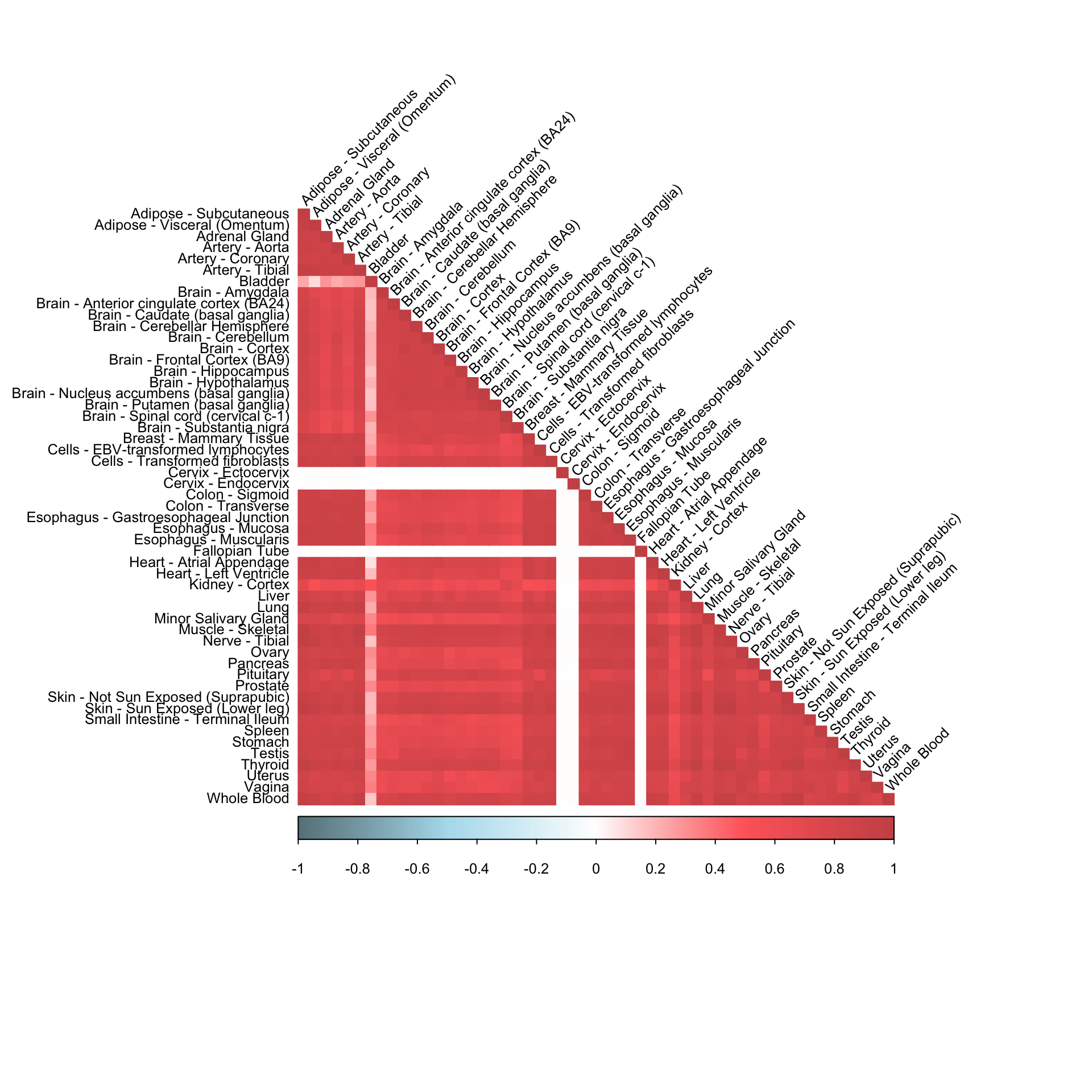
robospan[order(robospan, decreasing = T)[1:50]]ENSG00000134184 ENSG00000226278 ENSG00000184674 ENSG00000163682
0.7032426 0.6950521 0.6912778 0.6872412
ENSG00000233927 ENSG00000260246 ENSG00000204792 ENSG00000183298
0.6794906 0.6776832 0.6722359 0.6624388
ENSG00000196436 ENSG00000271581 ENSG00000235231 ENSG00000224114
0.6610709 0.6344899 0.6257342 0.6105988
ENSG00000198502 ENSG00000273295 ENSG00000152726 ENSG00000225760
0.6100595 0.6084643 0.5785107 0.5720591
ENSG00000237039 ENSG00000272004 ENSG00000227827 ENSG00000205571
0.5608029 0.5583093 0.5525935 0.5423600
ENSG00000272955 ENSG00000253570 ENSG00000214425 ENSG00000179344
0.5333256 0.5210327 0.5169869 0.5155531
ENSG00000262500 ENSG00000205578 ENSG00000254353 ENSG00000196656
0.5107885 0.5027289 0.5014584 0.4947770
ENSG00000213058 ENSG00000215513 ENSG00000225972 ENSG00000214110
0.4879866 0.4858357 0.4822919 0.4769688
ENSG00000226210 ENSG00000188234 ENSG00000236682 ENSG00000144115
0.4665672 0.4600686 0.4596699 0.4554153
ENSG00000214401 ENSG00000100376 ENSG00000226752 ENSG00000164308
0.4504711 0.4497744 0.4408376 0.4367149
ENSG00000096060 ENSG00000273018 ENSG00000228078 ENSG00000188933
0.4352561 0.4346918 0.4327234 0.4308759
ENSG00000187010 ENSG00000262539 ENSG00000013573 ENSG00000099984
0.4285536 0.4226383 0.4225531 0.4184471
ENSG00000204525 ENSG00000166435
0.4088639 0.3976043 corrplot(robocov_gtex[,,"ENSG00000179344"], diag = TRUE,
col = colorRampPalette(c("lightblue4", "lightblue2", "white", "indianred1", "indianred3"))(200),
tl.pos = "ld", tl.cex = 0.8, tl.col = "black",
rect.col = "white",na.label.col = "white",
method = "color", type = "lower", tl.srt=45)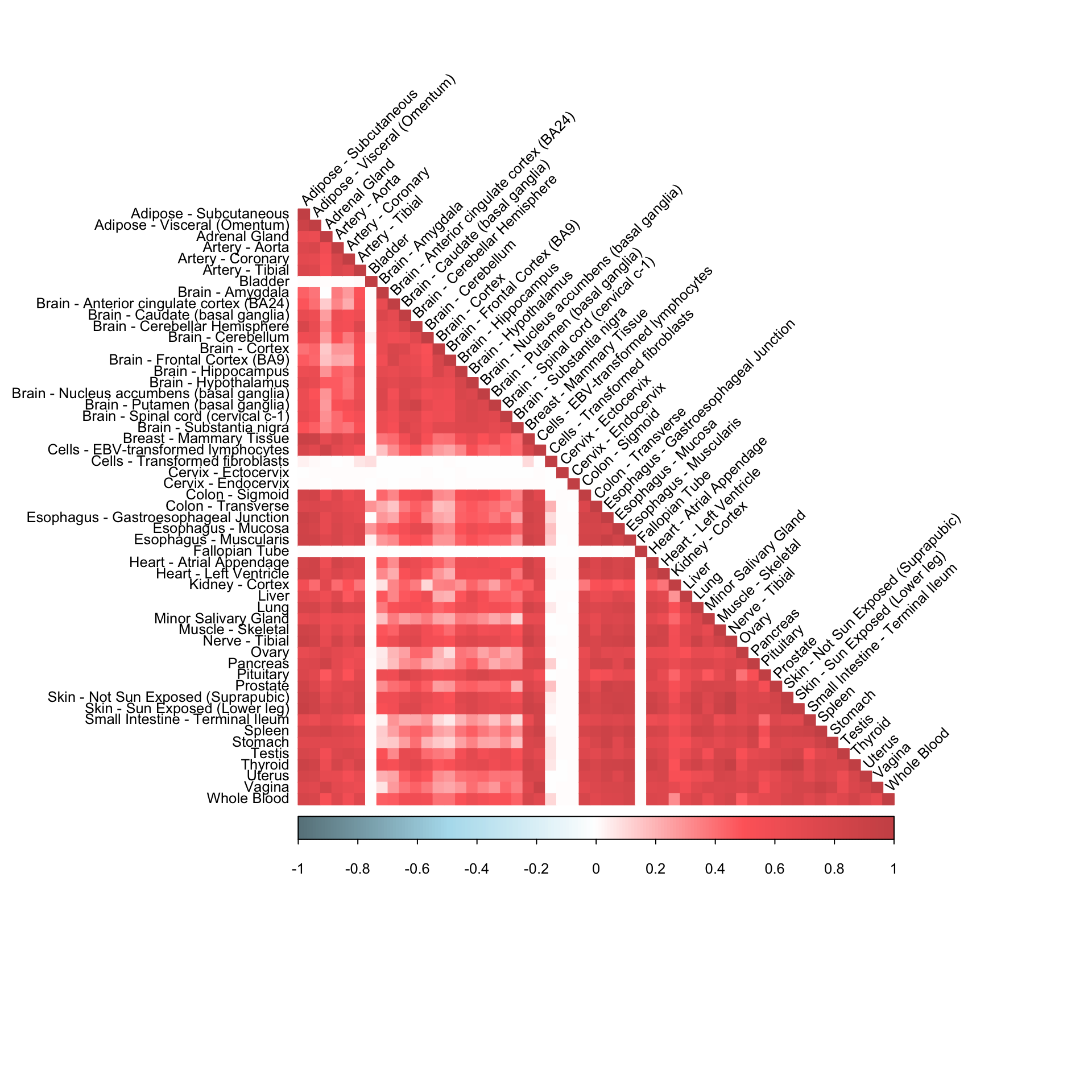
corrplot(robocov_gtex[,,"ENSG00000214425"], diag = TRUE,
col = colorRampPalette(c("lightblue4", "lightblue2", "white", "indianred1", "indianred3"))(200),
tl.pos = "ld", tl.cex = 0.8, tl.col = "black",
rect.col = "white",na.label.col = "white",
method = "color", type = "lower", tl.srt=45)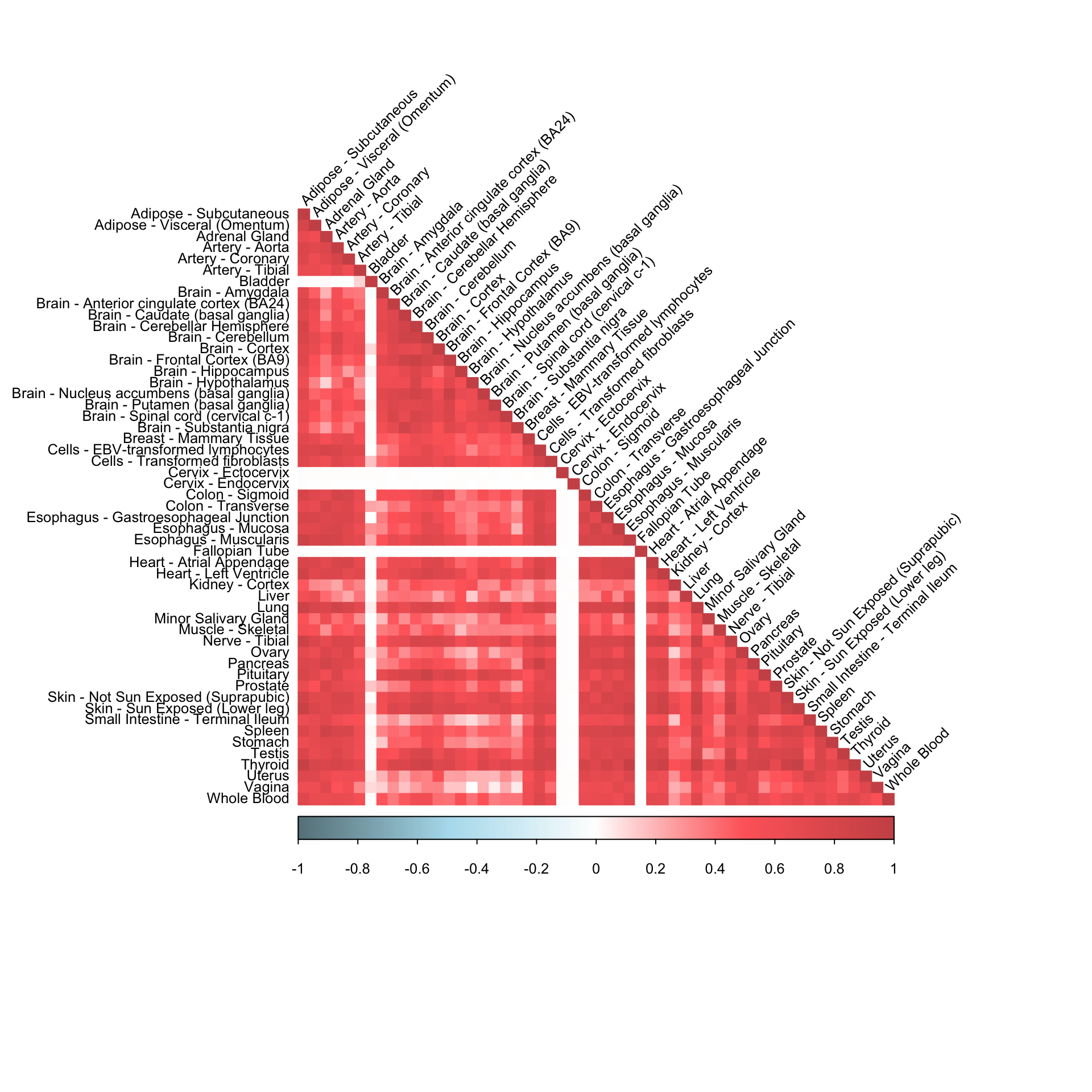
corrplot(robocov_gtex[,,"ENSG00000011600"], diag = TRUE,
col = colorRampPalette(c("lightblue4", "lightblue2", "white", "indianred1", "indianred3"))(200),
tl.pos = "ld", tl.cex = 0.8, tl.col = "black",
rect.col = "white",na.label.col = "white",
method = "color", type = "lower", tl.srt=45)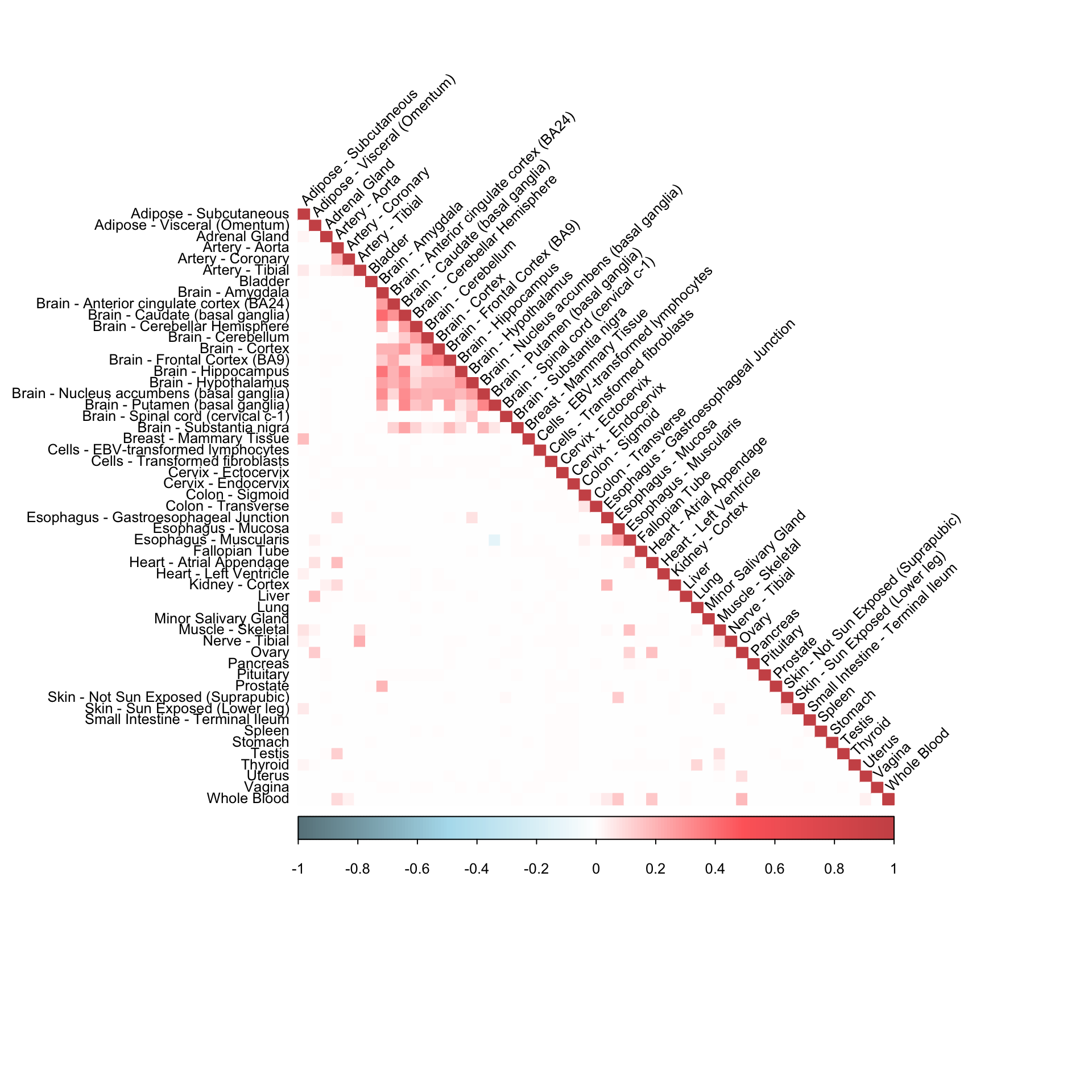
corrplot(robocov_gtex[,,"ENSG00000015475"], diag = TRUE,
col = colorRampPalette(c("lightblue4", "lightblue2", "white", "indianred1", "indianred3"))(200),
tl.pos = "ld", tl.cex = 0.8, tl.col = "black",
rect.col = "white",na.label.col = "white",
method = "color", type = "lower", tl.srt=45)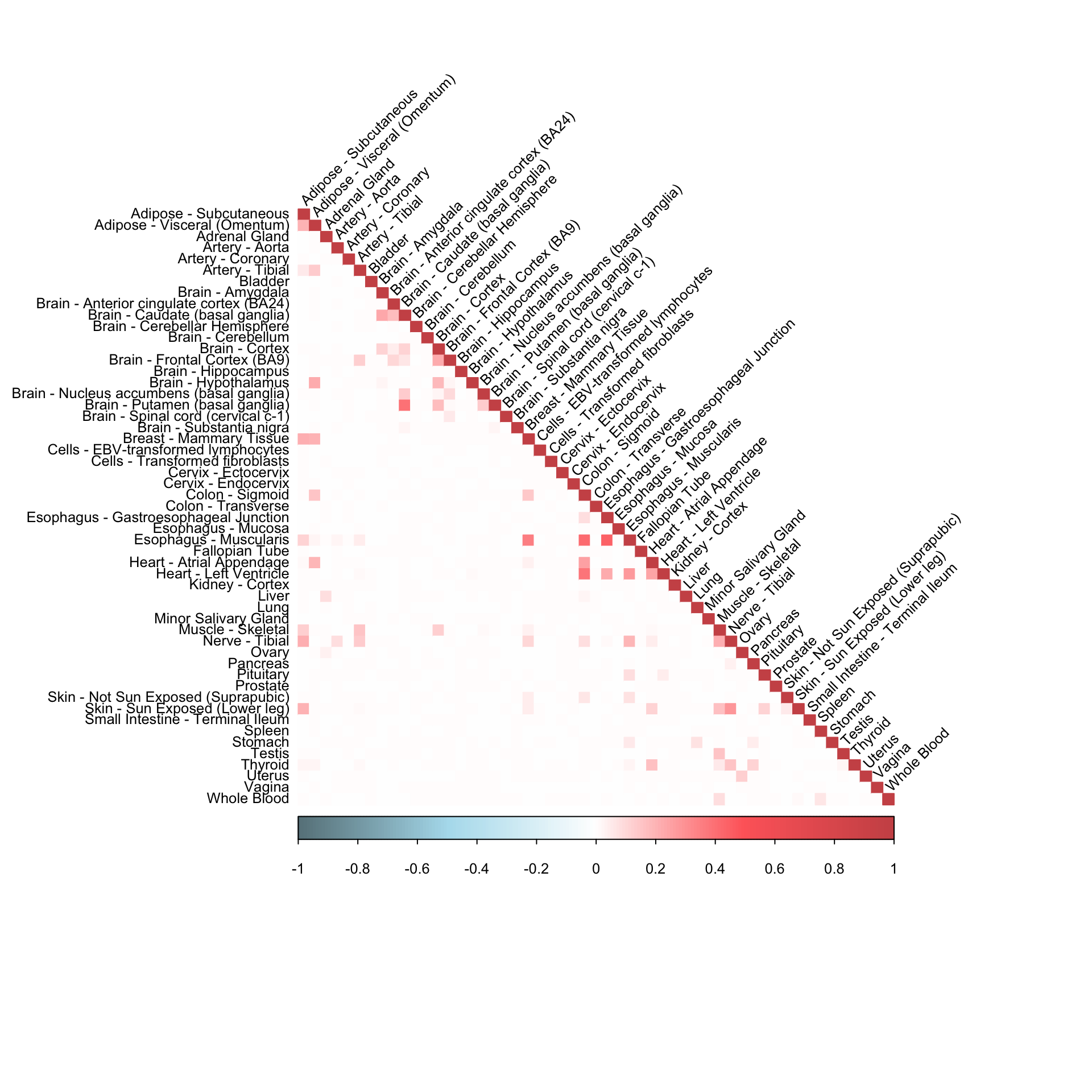
Session information
sessionInfo()R version 3.5.1 (2018-07-02)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS High Sierra 10.13.6
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] corrplot_0.84
loaded via a namespace (and not attached):
[1] workflowr_1.1.1 Rcpp_1.0.1 digest_0.6.19
[4] rprojroot_1.3-2 R.methodsS3_1.7.1 backports_1.1.4
[7] magrittr_1.5 git2r_0.23.0 evaluate_0.12
[10] stringi_1.4.3 whisker_0.3-2 R.oo_1.22.0
[13] R.utils_2.7.0 rmarkdown_1.10 tools_3.5.1
[16] stringr_1.4.0 yaml_2.2.0 compiler_3.5.1
[19] htmltools_0.3.6 knitr_1.20 This reproducible R Markdown analysis was created with workflowr 1.1.1